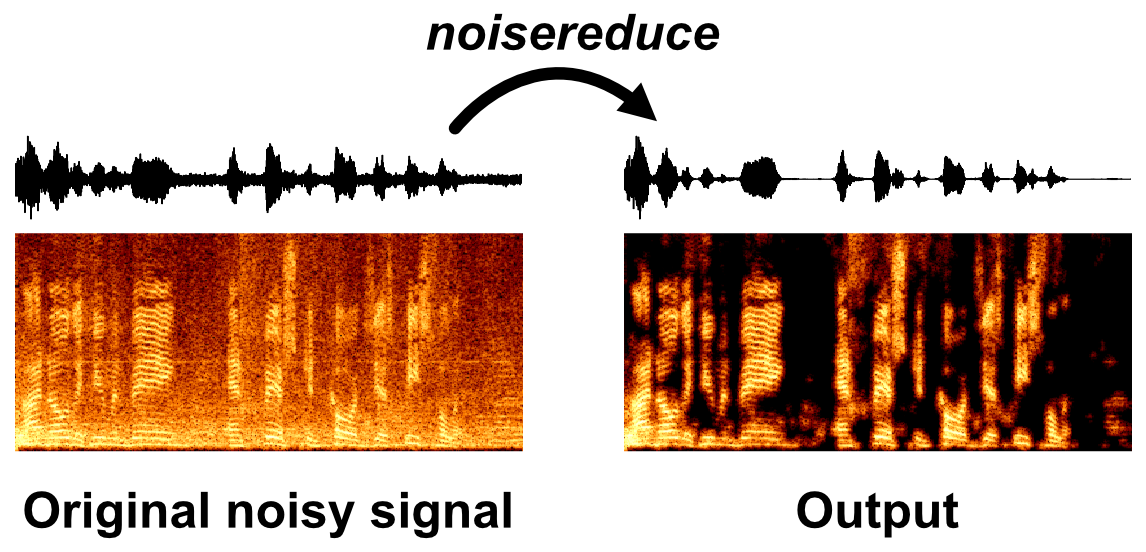
Noisereduce is a noise reduction algorithm in python that reduces noise in time-domain signals like speech, bioacoustics, and physiological signals. It relies on a method called "spectral gating" which is a form of Noise Gate. It works by computing a spectrogram of a signal (and optionally a noise signal) and estimating a noise threshold (or gate) for each frequency band of that signal/noise. That threshold is used to compute a mask, which gates noise below the frequency-varying threshold.
The most recent version of noisereduce comprises two algorithms:
- Stationary Noise Reduction: Keeps the estimated noise threshold at the same level across the whole signal
- Non-stationary Noise Reduction: Continuously updates the estimated noise threshold over time
- Added two forms of spectral gating noise reduction: stationary noise reduction, and non-stationary noise reduction.
- Added multiprocessing so you can perform noise reduction on bigger data.
- The new version breaks the API of the old version.
- The previous version is still available at
from noisereduce.noisereducev1 import reduce_noise - You can now create a noisereduce object which allows you to reduce noise on subsets of longer recordings
- The basic intuition is that statistics are calculated on each frequency channel to determine a noise gate. Then the gate is applied to the signal.
- This algorithm is based (but not completely reproducing) on the one outlined by Audacity for the noise reduction effect (Link to C++ code)
- The algorithm takes two inputs:
- A noise clip containing prototypical noise of clip (optional)
- A signal clip containing the signal and the noise intended to be removed
- A spectrogram is calculated over the noise audio clip
- Statistics are calculated over spectrogram of the the noise (in frequency)
- A threshold is calculated based upon the statistics of the noise (and the desired sensitivity of the algorithm)
- A spectrogram is calculated over the signal
- A mask is determined by comparing the signal spectrogram to the threshold
- The mask is smoothed with a filter over frequency and time
- The mask is appled to the spectrogram of the signal, and is inverted If the noise signal is not provided, the algorithm will treat the signal as the noise clip, which tends to work pretty well
- The non-stationary noise reduction algorithm is an extension of the stationary noise reduction algorithm, but allowing the noise gate to change over time.
- When you know the timescale that your signal occurs on (e.g. a bird call can be a few hundred milliseconds), you can set your noise threshold based on the assumption that events occuring on longer timescales are noise.
- This algorithm was motivated by a recent method in bioacoustics called Per-Channel Energy Normalization.
- A spectrogram is calculated over the signal
- A time-smoothed version of the spectrogram is computed using an IIR filter aplied forward and backward on each frequency channel.
- A mask is computed based on that time-smoothed spectrogram
- The mask is smoothed with a filter over frequency and time
- The mask is appled to the spectrogram of the signal, and is inverted
pip install noisereduce
from scipy.io import wavfile
import noisereduce as nr
# load data
rate, data = wavfile.read("mywav.wav")
# perform noise reduction
reduced_noise = nr.reduce_noise(y=data, sr=rate)
wavefile.write("mywav_reduced_noise.wav", rate, reduced_noise)
y : np.ndarray [shape=(# frames,) or (# channels, # frames)], real-valued
input signal
sr : int
sample rate of input signal / noise signal
y_noise : np.ndarray [shape=(# frames,) or (# channels, # frames)], real-valued
noise signal to compute statistics over (only for non-stationary noise reduction).
stationary : bool, optional
Whether to perform stationary, or non-stationary noise reduction, by default False
prop_decrease : float, optional
The proportion to reduce the noise by (1.0 = 100%), by default 1.0
time_constant_s : float, optional
The time constant, in seconds, to compute the noise floor in the non-stationary
algorithm, by default 2.0
freq_mask_smooth_hz : int, optional
The frequency range to smooth the mask over in Hz, by default 500
time_mask_smooth_ms : int, optional
The time range to smooth the mask over in milliseconds, by default 50
thresh_n_mult_nonstationary : int, optional
Only used in nonstationary noise reduction., by default 1
sigmoid_slope_nonstationary : int, optional
Only used in nonstationary noise reduction., by default 10
n_std_thresh_stationary : int, optional
Number of standard deviations above mean to place the threshold between
signal and noise., by default 1.5
tmp_folder : [type], optional
Temp folder to write waveform to during parallel processing. Defaults to
default temp folder for python., by default None
chunk_size : int, optional
Size of signal chunks to reduce noise over. Larger sizes
will take more space in memory, smaller sizes can take longer to compute.
, by default 60000
padding : int, optional
How much to pad each chunk of signal by. Larger pads are
needed for larger time constants., by default 30000
n_fft : int, optional
length of the windowed signal after padding with zeros.
The number of rows in the STFT matrix ``D`` is ``(1 + n_fft/2)``.
The default value, ``n_fft=2048`` samples, corresponds to a physical
duration of 93 milliseconds at a sample rate of 22050 Hz, i.e. the
default sample rate in librosa. This value is well adapted for music
signals. However, in speech processing, the recommended value is 512,
corresponding to 23 milliseconds at a sample rate of 22050 Hz.
In any case, we recommend setting ``n_fft`` to a power of two for
optimizing the speed of the fast Fourier transform (FFT) algorithm., by default 1024
win_length : [type], optional
Each frame of audio is windowed by ``window`` of length ``win_length``
and then padded with zeros to match ``n_fft``.
Smaller values improve the temporal resolution of the STFT (i.e. the
ability to discriminate impulses that are closely spaced in time)
at the expense of frequency resolution (i.e. the ability to discriminate
pure tones that are closely spaced in frequency). This effect is known
as the time-frequency localization trade-off and needs to be adjusted
according to the properties of the input signal ``y``.
If unspecified, defaults to ``win_length = n_fft``., by default None
hop_length : [type], optional
number of audio samples between adjacent STFT columns.
Smaller values increase the number of columns in ``D`` without
affecting the frequency resolution of the STFT.
If unspecified, defaults to ``win_length // 4`` (see below)., by default None
n_jobs : int, optional
Number of parallel jobs to run. Set at -1 to use all CPU cores, by default 1
If you use this code in your research, please cite it:
@software{tim_sainburg_2019_3243139,
author = {Tim Sainburg},
title = {timsainb/noisereduce: v1.0},
month = jun,
year = 2019,
publisher = {Zenodo},
version = {db94fe2},
doi = {10.5281/zenodo.3243139},
url = {https://doi.org/10.5281/zenodo.3243139}
}
@article{sainburg2020finding,
title={Finding, visualizing, and quantifying latent structure across diverse animal vocal repertoires},
author={Sainburg, Tim and Thielk, Marvin and Gentner, Timothy Q},
journal={PLoS computational biology},
volume={16},
number={10},
pages={e1008228},
year={2020},
publisher={Public Library of Science}
}
Project based on the cookiecutter data science project template. #cookiecutterdatascience