This implements a method to apply Topological Data Analysis (TDA) as a classifier.
The overall idea is to use variable-sized neighborhoods to perform the classification instead of using a fixed collection of points as the kNN family of methods does.
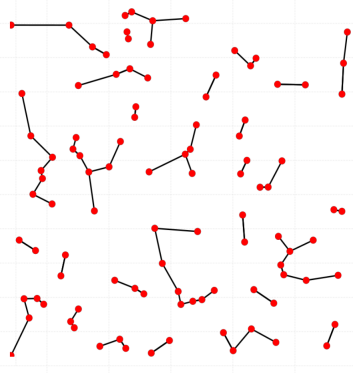
When you connect each point of your point cloud with its k nearest-neighbors, you will get a nearest-neighbors graph, as we show in the figure:
From a mathematical point of view, we know that a Graph is a 1-dimensional simplicial complex. A Graph is also the
1-skeleton of any simplicial complex. The k-NN neighborhoods are a collection of
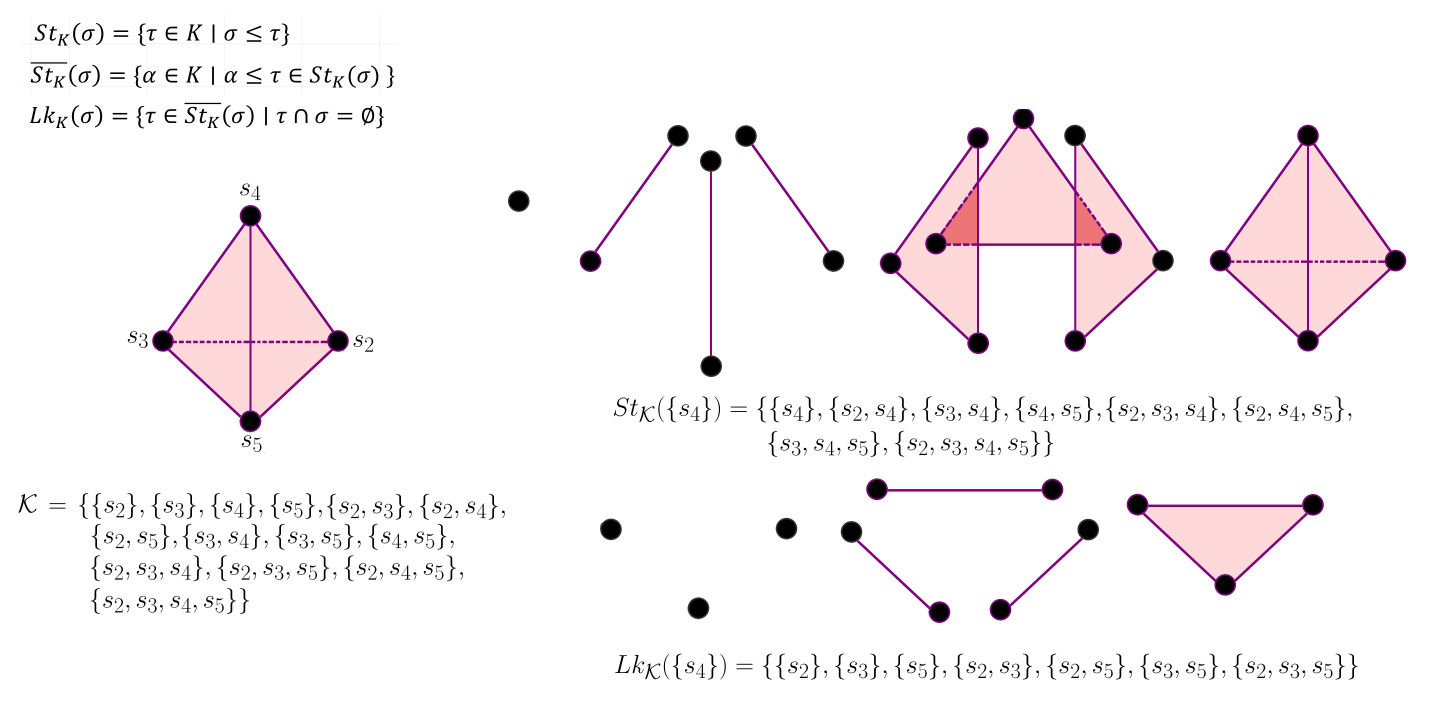
The star and link of simplices give us the simplicial neighborhoods of a simplex in a simplicial complex. As we know, in
our dataset, we have some labeled points and unlabeled the remaining ones. So we need to figure out how to propagate labels from
labeled to unlabeled points. We can consider simplices as relationships between points at different orders. So a 1-simplex
(edge) means the classical relationship between two points, a 2-simplex (triangle), a relationship between three points,
and so on. With high dimensional simplices, we get high order relationships between points.
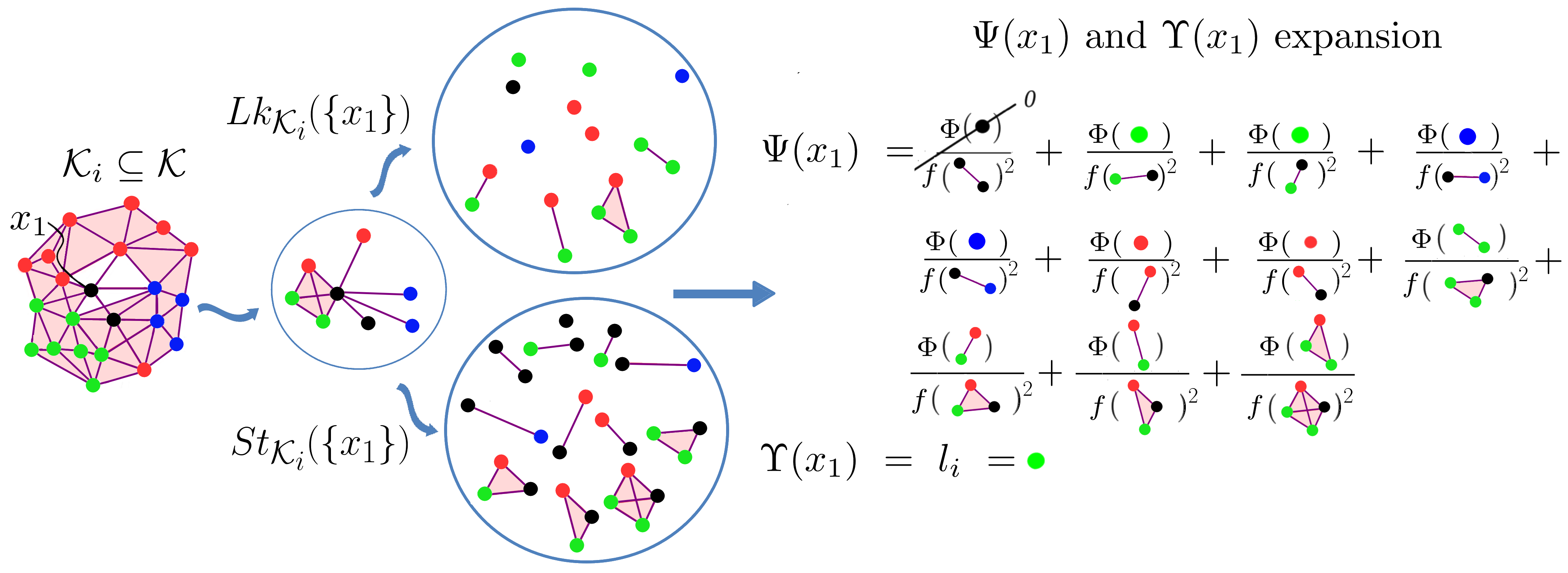
Thus, to label a point
This solution enriches the information we can analyze from our data and gives us more insight. However,
a big issue remains, how can we be sure that our simplicial complex represents our data? This question arises from the fact that we can build a high number of simplicial complexes from the same point cloud.
Even with the chosen type of simplicial complex
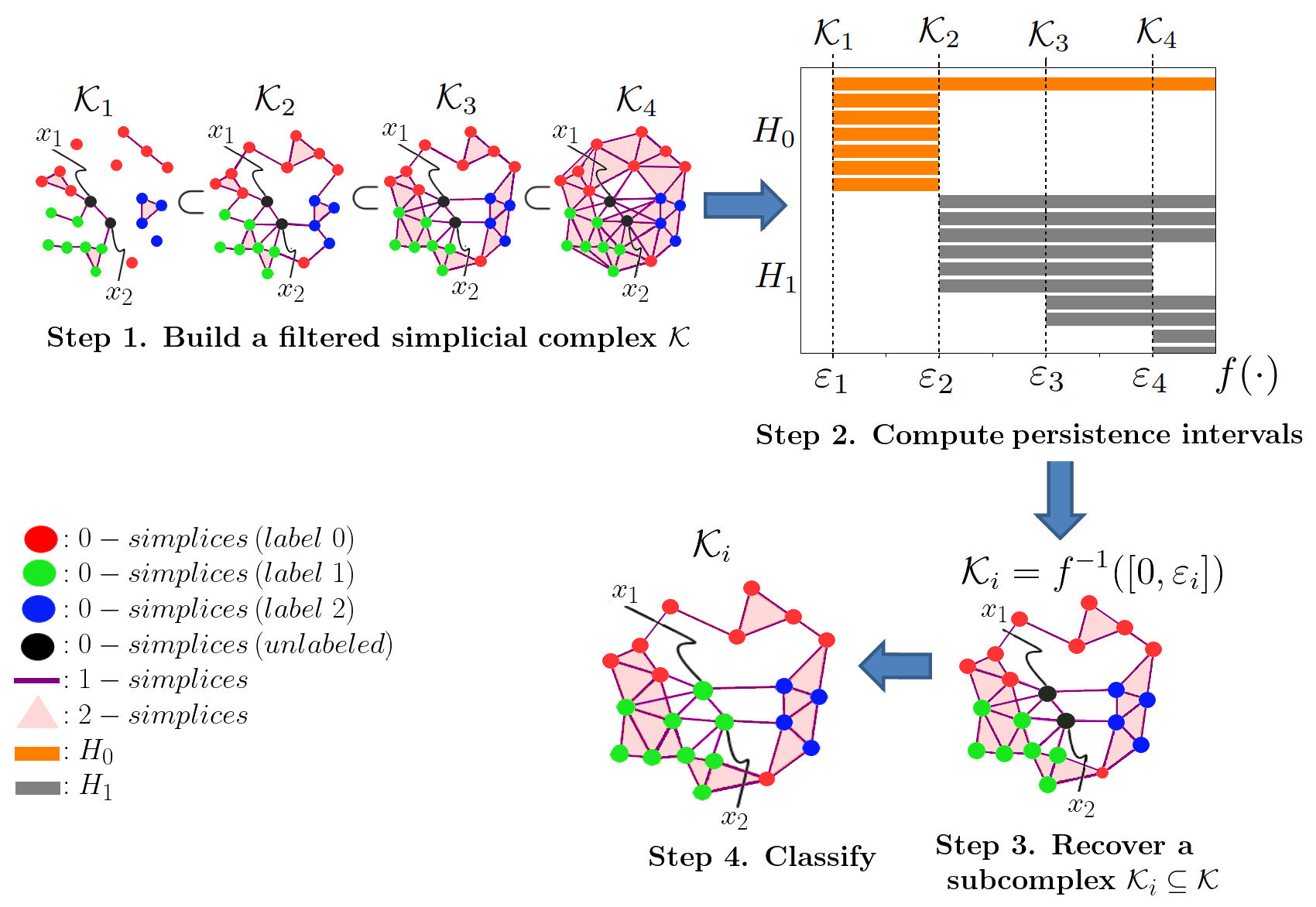
This is an old problem that has been studied in TDA. The typical approach is to forget any specific threshold and work with all of them up to a maximum defined threshold value. The idea is to explore every simplicial complex in each scale of the threshold value. We get a nested and increased collection of simplicial complexes containing the previous ones called a Filtration. Then we can use the TDA working horse "Persistent Homology (PH)," a technique capable of capturing and understanding the evolution of topological features conforming to the threshold value is increased. The process we followed here was to use PH to understand our dataset topologically, taking advantage of that information to extract a simplicial complex from the filtration, which approximates our dataset well enough. Then, use the label propagation method as mentioned earlier in that sub-complex.
The overall process is summarized in the Figure:
There is a req.txt file automatically generated but it can be installed manually. We also provide a ipynb file to run.
- Install Anaconda
- make a new environment
conda create --name tdabc_env
- Install gudhi according to (https://gudhi.inria.fr/python/latest/installation.html). We choose the conda instalation way:
conda install -c conda-forge gudhi
- Install scikit-learn
conda install -c conda-forge scikit-learn
- Install matplotlib
conda install -c conda-forge matplotlib
to work with latex
sudo apt install texlive-fonts-recommended texlive-fonts-extra
sudo apt install dvipng
- Install umap (optional). In case your want to install it, you need to downgrade your python to python 3.8, and every package as well to be compatible with this version.
conda install -c conda-forge umap-learn pip install umap-learn
- Install h5py
conda install -c conda-forge h5py
the entry point is the main.py
to run the project it is enough with:
python main.py
however the main accepts arguments to configure the execution:
usage: main.py [-h] [-o OPTION] [-d DATASET] [-path PATH] [-i ITERATION] [-n NUMBER_DATASETS] [-m METRIC] [-t TRANSFORMATION]
Where:
-> -o --option help="Select option to execute 1: Multi KRNN experiment, 2: KRNN, 3: Conventional experiment " type=int
-> -d --dataset help="Select the datasets" type=int
-> -path --path help="Define folder to save results" type=str
-> -i --iteration" help="Iteration number for option=1" type=int
-> -n --number_datasets help="Number of datasets for option=1 type=int
-> -m --metric help="Metric to build the simplicial comples. A collection of possible metrics are: ['cityblock', 'cosine', " "'euclidean', 'l1', 'l2', 'manhattan', 'nan_euclidean','braycurtis'," " 'canberra', 'chebyshev','correlation', 'dice', 'hamming', 'jaccard'," " 'kulsinski', 'mahalanobis', 'minkowski', 'rogerstanimoto', " "'russellrao', 'seuclidean', 'sokalmichener', " "'sokalsneath', 'sqeuclidean', 'yule']" type=str
-> -t --transformation help="Data Transformation type to apply, types are: 0=NONE, SIMPLE=1, SQUARED=2, NLOG=3, NORM=4, INV=5, ELOG=6, REDUCTION=7" type=int
When select a data transformation of reduction, it means that you want to perform a dimensionality reduction, we implement three algorithms PCA=0, TSNE=1, and UMAP=2 there is not interface to access to those method from the entry point.
- To run the experiment in the eight datasets:
python main.py -o 3 -p YOUR_RESULT_PATH
you can also define -m YOUR_DESIRED_METRIC but it is optional because the euclidean distance is default.
- To run the experiment of generating the results of all datasets (Figure 7 in the paper)
python main.py -o 1 -i 1 -p "./docs/ADAC_TDABC_RESULTS"
This option 1 what does is to collect all dataset info inside the path directory and looking for *metrics.txt files which are where all metric results are stored. Note that this method, assumes all datasets folder has the same number of experiements.
To generate the RKNN experiments with 16 variations of samples, we need to provide the folder where all KRNN datasets are stored. The algorithms detects by itself according to the structure of data if its the case of datasets or the KRNN one.
To RUN the RKNN experiment, we must specify the iterations that we need to consider.
python main.py -o 1 -i 3 -n 16 -p "./docs/KRNN_EXPERIMENTS"
Generates n datasets 50:50, 50:100, 50:150, ..., 50:800 and executes i iterations of each one of our TDABC and also generate the plots.
NOTE that both experiments with option -o 1 are mutually excluyent and they should not run on the same directory
Any comment, please do not hesitate to write a mail to Rolando Kindelan Nuñez ( rolan2kn.at.gmail.com , rkindela.at.dcc.uchile.cl), twitter: at.rolan2kn.
Kindelan, Rolando., et al. “A Topological Data Analysis Based Classifier.” ArXiv.org, February 3rd, 2021, arxiv.org/abs/2111.05214.
@misc{rol2021topological, title={A Topological Data Analysis Based Classifier}, author={Rolando Kindelan and José Frías and Mauricio Cerda and Nancy Hitschfeld}, year={2021}, eprint={2111.05214}, archivePrefix={arXiv}, primaryClass={cs.LG} }