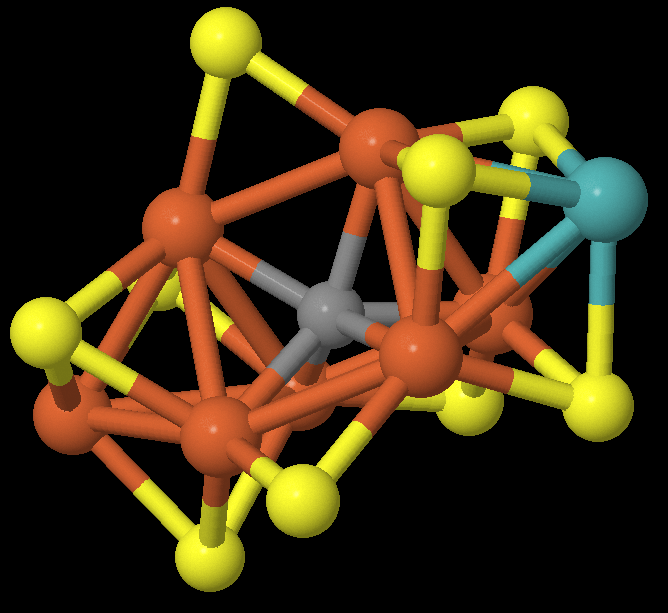
This is the project that Minsik Cho and I worked on during a mentorship program
hosted by the Quantum Open Software Foundation (https://qosf.org), over the
winter of the year 2020. Our project attempts to apply the Variational Quantum
Eigensolver -- an algorithm to estimate energy levels of molecules -- to the
FeMo1 cofactor of nitrogenase. This cluster is responsible for
nitrogenase's remarkable ability to break the triple bond of the N_2 molecule
at atmospheric temperatures and pressures, and the ability to calculate energies
for chemical intermediates of the cluster is critical to understanding the
viable reaction pathways.
This is ongoing work, and may be in various states of disarray... be warned.
Visit our Project Webpage for more details.
examples/: Example scriptssrc/: Larger proof-of-concept scripts (less stable)lib/: Modified qiskit codetests/: Unit tests. Run withpytest tests/hdf5_files/: SCF results. Load withQMolecule.load()molecules/: Molecular geometries (pulled from the Protein Data Bank)
- Install conda.
- Clone this repository and
cdto it. - Create the environment using:
conda env create -f environment.yml - Activate the conda environment:
conda activate qosf - Run the test suite:
pytest tests/ - Run the scripts from the repository root (e.g.):
python examples/gen_qubit_op_lih.py python src/gen_hamiltonian_qiskit.py
Special thanks to Vesselin for providing guidance throughout this project, Michał Stęchły for organizing the mentorship program, and the rest of the folks at the Quantum Open Software Foundation for their work in enabling free and open-source research for quantum computing.
Questions? Reach us at:
- Roy Tu (kroytu [at] berkeley [dot] edu)
- Minsik Cho