This repository contains an R package for visualizing data from the NCEI/WDS Global Significant Earthquake Database. The main functions it provides are for adding layers to ggplot where you can make a timeline showing the significant earthquakes across a given time period, and for generating leaflet maps showing the epicenters for significant earthquake events.
You can install this R package from GitHub using devtools:
library(devtools)
install_github("rizem2/noaa-earthquakes")Loading the relevant packages for these examples:
library(earthquakes)
library(tidyverse)
library(leaflet)The functions provided with this package fall into 3 general categories: reading/cleaning data, creating ggplot2 layers, and generating leaflet maps.
This package comes with the raw data file
earthquakes-2021-03-08_17-02-58_-0500.tsv taken directly from the NCEI
database API along with three functions for reading and cleaning the raw
data:
eq_read_data- reads a raw data file into a data frame and formats some of the columns. It takes a filepath as an argument indicating the raw data file to be read into R. If called with no arguments, it defaults to the supplied .tsv file.eq_location_clean- splits theLOCATIONcolumn into aCOUNTRYandREGIONcolumn. Takes a dataframe as an argument, presumably the output fromeq_read_data. If called with no arguments, callseq_read_data.eq_clean_data- uses the rawYEAR,MONTH,DAYcolumns to form aDATEcolumn using lubridate. Takes a dataframe as an argument, presumably the output fromeq_location_clean. If called with no arguments, callseq_location_cleanand returns a fully “cleaned” dataframe of the supplied raw data.
Viewing the 10 most deadly earthquakes since the year 2000:
data <- eq_clean_data() %>%
dplyr::filter(!is.na(TOTAL_DEATHS)) %>%
dplyr::select(DATE, YEAR, COUNTRY, REGION, LONGITUDE, LATITUDE, MAG, TOTAL_DEATHS) %>%
tidyr::drop_na()
data %>%
dplyr::filter(YEAR > 1999) %>%
dplyr::arrange(desc(TOTAL_DEATHS)) %>%
head(n = 10)
#> # A tibble: 10 x 8
#> DATE YEAR COUNTRY REGION LONGITUDE LATITUDE MAG TOTAL_DEATHS
#> <date> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 2010-01-12 2010 HAITI PORT-AU-PRIN~ -72.5 18.5 7 316000
#> 2 2004-12-26 2004 INDONES~ SUMATRA: AC~ 95.9 3.32 9.1 227899
#> 3 2008-05-12 2008 CHINA SICHUAN PROV~ 103. 31.0 7.9 87652
#> 4 2005-10-08 2005 PAKISTAN MUZAFFARABAD~ 73.6 34.5 7.6 76213
#> 5 2003-12-26 2003 IRAN SOUTHEASTERN~ 58.3 29.0 6.6 31000
#> 6 2001-01-26 2001 INDIA GUJARAT: BH~ 70.2 23.4 7.7 20005
#> 7 2011-03-11 2011 JAPAN HONSHU 142. 38.3 9.1 18429
#> 8 2015-04-25 2015 NEPAL KATHMANDU; I~ 84.7 28.2 7.8 8200
#> 9 2006-05-26 2006 INDONES~ JAVA: BANTU~ 110. -7.96 6.3 5749
#> 10 2018-09-28 2018 INDONES~ SULAWESI 120. -0.178 7.5 4340The next two functions are used to add custom layers to ggplot
This package comes with a custom ggplot2 Geom GeomTimeline for
visualizing Earthquake characteristics (such as Magnitude, Death Toll,
Damages, etc.) on a timeline. To use this Geom, add a ggplot2 layer
using the function geom_timeline.
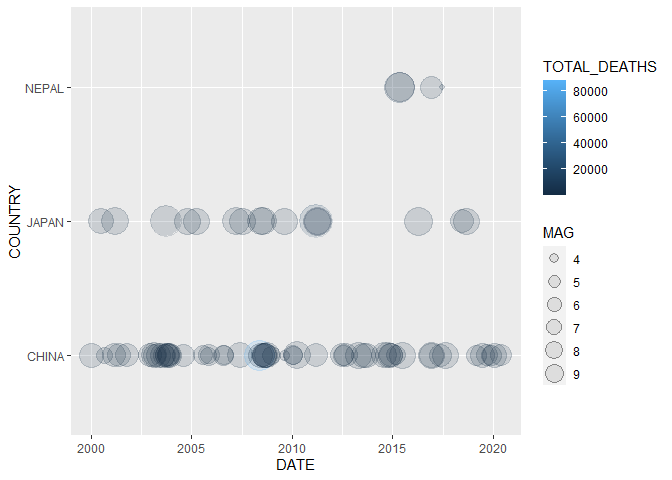
Plot a timeline of deadly earthquakes in 3 different countries:
data %>% dplyr::filter(YEAR > 1999,
COUNTRY %in% c("JAPAN", "CHINA","NEPAL")) %>%
ggplot2::ggplot() +
geom_timeline(aes(x = DATE,
y = COUNTRY,
size = MAG,
color = TOTAL_DEATHS))We can extend these visualizations even further by adding labels to our
timelines using the GeomTimelineLabel Geom. Simply add another ggplot2
layer using the geom_timeline_label in conjunction with
geom_timeline.
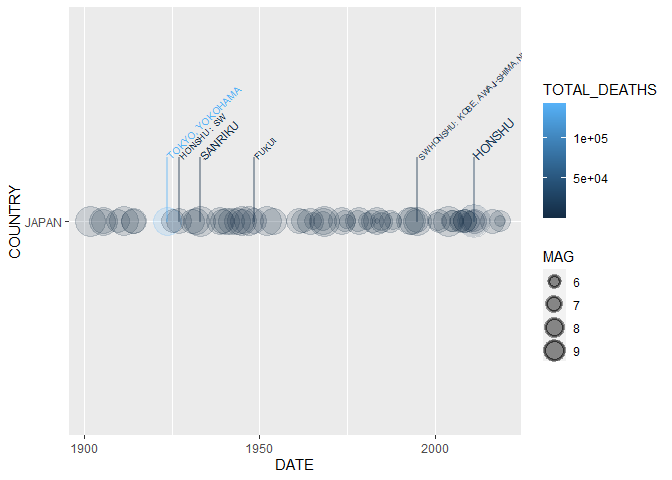
Labeling the 5 most deadly earthquakes (6 are shown since 5th is “tied”)
in the Japan since 1900, note that the column we use for labeling is
indicated by the mag aesthetic:
data %>% dplyr::filter(COUNTRY == "JAPAN",
YEAR >= 1900) %>%
ggplot2::ggplot(aes(x = DATE,
y = COUNTRY,
size = MAG,
color = TOTAL_DEATHS,
label = REGION,
mag = TOTAL_DEATHS)) +
geom_timeline() +
geom_timeline_label(aes(n_max = 5))The final two functions generate leaflet maps for visualizing earthquake
epicenters on actual maps. The first function eq_map generates a
leaflet map from a dataframe with columns LONGITUDE, LATITUDE, and
MAG. eq_map takes one argument annot_col which denotes the column
of the dataframe to be used for annotation, by default uses DATE.
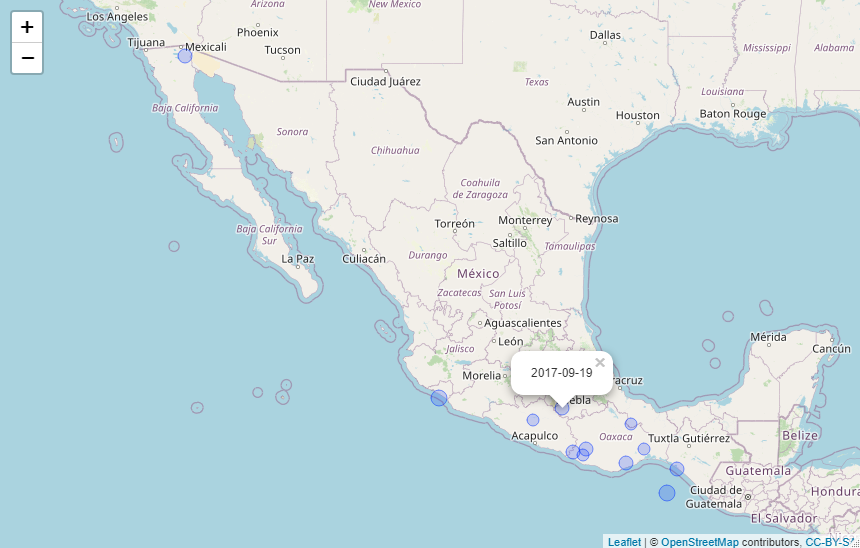
Plot the epicenters on a leaflet map:
data %>%
dplyr::filter(COUNTRY == "MEXICO",
YEAR >= 2000) %>%
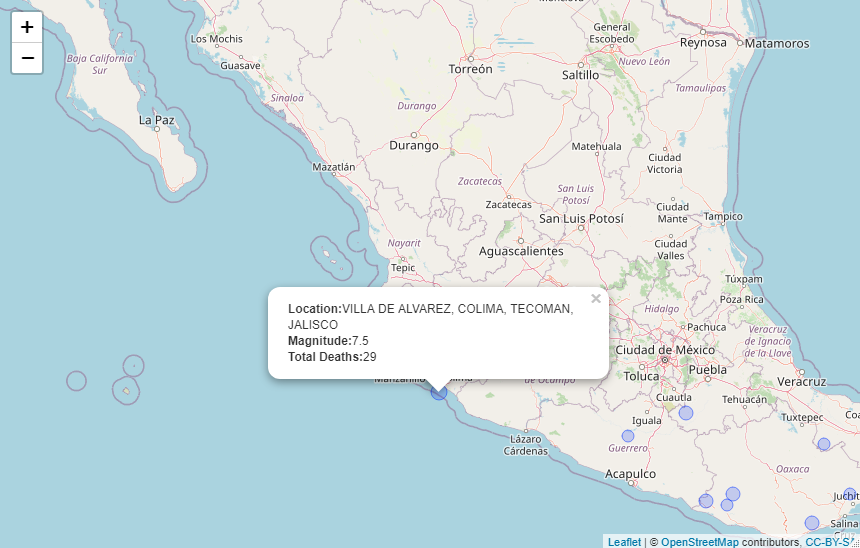
eq_map(annot_col = "DATE")The function eq_create_label generates labels from the input
dataframe. We can use eq_create_label within dplyr::mutate to modify
the input dataframe by creating a column to store the desired pop-up
text as an HTML string. The generated pop-ups will display the location,
magnitude and total death count (if available) for any given earthquake.
data %>%
dplyr::filter(COUNTRY == "MEXICO",
YEAR >= 2000) %>%
dplyr::mutate(popup_text = eq_create_label(.)) %>%
eq_map(annot_col = "popup_text")