This is an older copy of https://github.com/VLL-HD/FrEIA, used within BARC.
This is the Framework for Easily Invertible Architectures (FrEIA).
- Construct Invertible Neural Networks (INNs) from simple invertible building blocks.
- Quickly construct complex invertible computation graphs and INN topologies.
- Forward and inverse computation guaranteed to work automatically.
- Most common invertible transforms and operations are provided.
- Easily add your own invertible transforms.
Table of contents
Our following papers use FrEIA, with links to code given below.
"Generative Classifiers as a Basis for Trustworthy Image Classification" (CVPR 2021)
"Training Normalizing Flows with the Information Bottleneck for Competitive Generative Classification" (Neurips 2020)
- Paper: https://arxiv.org/abs/2001.06448
- Code: https://github.com/VLL-HD/exact_information_bottleneck
"Disentanglement by Nonlinear ICA with General Incompressible-flow Networks (GIN)" (ICLR 2020)
"Guided Image Generation with Conditional Invertible Neural Networks" (2019)
- Paper: https://arxiv.org/abs/1907.02392
- Supplement: https://drive.google.com/file/d/1_OoiIGhLeVJGaZFeBt0OWOq8ZCtiI7li
- Code: https://github.com/VLL-HD/conditional_invertible_neural_networks
"Analyzing inverse problems with invertible neural networks." (ICLR 2019)
| Package | Version |
| Python | >= 3.7 |
| Pytorch | >= 1.0.0 |
| Numpy | >= 1.15.0 |
To just install the framework:
pip install git+https://github.com/VLL-HD/FrEIA.gitFor development:
# first clone the repository
git clone https://github.com/VLL-HD/FrEIA.git
# then install in development mode, so that changes don't require a reinstall
cd FrEIA
python setup.py developBesides the general tutorial below, the complete documentation is found in
the ./docs/ directory, or at
https://vll-hd.github.io/FrEIA
To jump straight into the code, see this basic usage example, which learns and then samples from the moons dataset provided in sklearn. For information on the structure of FrEIA, as well as more detailed examples of networks, including custom invertible operations, see the full tutorial below.
# standard imports
import torch
import torch.nn as nn
from sklearn.datasets import make_moons
# FrEIA imports
import FrEIA.framework as Ff
import FrEIA.modules as Fm
BATCHSIZE = 100
N_DIM = 2
# we define a subnet for use inside an affine coupling block
# for more detailed information see the full tutorial
def subnet_fc(dims_in, dims_out):
return nn.Sequential(nn.Linear(dims_in, 512), nn.ReLU(),
nn.Linear(512, dims_out))
# a simple chain of operations is collected by ReversibleSequential
inn = Ff.SequenceINN(N_DIM)
for k in range(8):
inn.append(Fm.AllInOneBlock, subnet_constructor=subnet_fc, permute_soft=True)
optimizer = torch.optim.Adam(inn.parameters(), lr=0.001)
# a very basic training loop
for i in range(1000):
optimizer.zero_grad()
# sample data from the moons distribution
data, label = make_moons(n_samples=BATCHSIZE, noise=0.05)
x = torch.Tensor(data)
# pass to INN and get transformed variable z and log Jacobian determinant
z, log_jac_det = inn(x)
# calculate the negative log-likelihood of the model with a standard normal prior
loss = 0.5*torch.sum(z**2, 1) - log_jac_det
loss = loss.mean() / N_DIM
# backpropagate and update the weights
loss.backward()
optimizer.step()
# sample from the INN by sampling from a standard normal and transforming
# it in the reverse direction
z = torch.randn(BATCHSIZE, N_DIM)
samples, _ = inn(z, rev=True)"Why does FrEIA even exist? RealNVP can be implemented in ~100 lines of code!"
That is correct, but the concept of INNs is more general: For any computation graph, as long as each node in the graph is invertible, and there are no loose ends, the entire computation is invertible. This is also true if the operation nodes have multiple in- or outputs, e.g. concatenation (n inputs, 1 output). So we need a framework that allows to define an arbitrary computation graph, consisiting of invertible operations.
For example, consider wanting to implement some complicated new INN
architecture, with multiple in- and outputs, skip connections, a conditional part, ...:
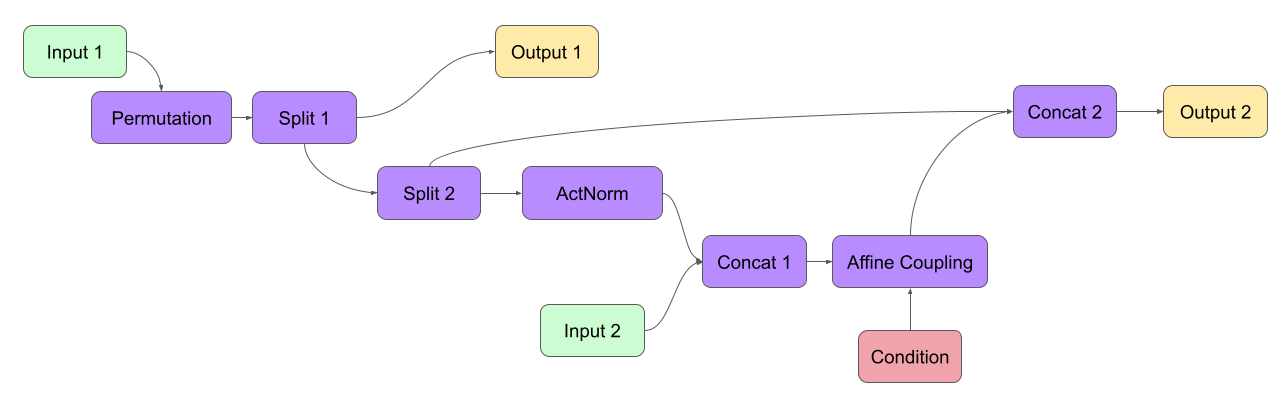
To allow efficient prototyping and experimentation with such architectures, we need a framework that can perform the following tasks:
- As the inputs of operations depend on the outputs of others, we have to infer the order of operations, both for the forward and the inverse direction.
- The operators have to be initialized with the correct input- and output sizes in mind (e.g. required number of weights), i.e. we have to perform shape inference on the computation graph.
- During the computation, we have to keep track of intermediate results (edges in the graph) and store them until they are needed.
- We want to use pytorch methods and tools, such as
.cuda(),.state_dict(),DataParallel(), etc. on the entire computation graph, without worrying whether they work correctly or having to fix them.
Along with an interface to define INN computation graphs and invertible
operators within, these are the main tasks that FrEIA addresses.
The building blocks of the INN computation graph are the nodes in it.
They are provided through the FrEIA.framework.Node class.
The computation graph is constructed by constructing each node, given its
inputs (defining one direction of the INN as the 'forward' computation).
More specifically:
The
Node-subclassInputNoderepresents an input to the INN, and its constructor only takes the dimensions of the data (except the batch dimension). E.g. for a 32x32 RGB image:in1 = InputNode(3, 32, 32, name='Input 1')
The
nameargument can be omitted in principle, but it is recommended in general, as it appears e.g. in error messages.Each
Node(and derived classes) has propertiesnode.out0,node.out1, etc., depending on its number of outputs. Instead ofnode.out{i}, it is equivalent to use a tuple(node, i), which is useful if you e.g. want to loop over 10 outputs of a node.Each
Nodeis initialized given a list of its inputs as the first constructor argument, along with other arguments covered later (omitted as '...' in the following, in particular defining what operation the node should represent). For Permutation in the example above, this would look like the this:perm = Node([in1.out0], ..., name='Permutation')
Or for Merge 2:
merge2 = Node([affine.out0, split2.out1], ..., name='Merge 2')
Conditions are passed as a list through the
conditionsargument:affine = Node([merge1.out0], ..., conditions=[cond], name='Affine Coupling')
The
Node-subclassOutputNodeis used for the outputs. The INN as a whole will return the result at this node.Conditions (as in the cINN paper) are represented by
ConditionNode, whose constructor is identical to theInputNode.Take note of several features for convenience (also see examples below): 1.) If a preceding node only has a single output, it is also equivalent to directly use
nodeinstead ofnode.out0in the constructor of following nodes. 2.) If a node only takes a sinlge input/condition, you can directly use only that input in the constructor instead of a list, i.e.node.out0instead of[node.out0].From the list of nodes, the INN is represented by the class
FrEIA.framework.GraphINN. The constructor takes a list of all the nodes in the INN (order irrelevant).The
GraphINNis a subclass oftorch.nn.Module, and can be used like any other torchModule. For the computation, the inputs are given as a list of torch tensors, or just a single torch tensor if there is only one input. To perform the inverse pass, therevargument has to be set toTrue(see examples).
Above, we only covered the construction of the computation graph itself, but so
far we have not shown how to define the operations represented by each node.
Therefore, we will take a closer look at the Node constructor and its
arguments:
Node(inputs, module_type, module_args, conditions=[], name=None)The arguments of the Node constructor are the following:
inputs: A list of outputs of other nodes, that are used as inputs for this node (discussed above)module_type: This argument gives the class of operation to be performed by this node, for exampleGLOWCouplingBlockfor a coupling block following the GLOW-design. Many implemented classes can be found in the documentation under https://vll-hd.github.io/FrEIA/modules/index.htmlmodule_args: This argument is a dictionary. It provides arguments for themodule_type-constructor. For instance, a random invertible permutation (module_type=PermuteRandom) can accept the argumentseed, so we could usemodule_args={'seed': 111}. If no arguments are specified we must pass an empty dictionary{}.
All coupling blocks (GLOW, RNVP, NICE), merit special discussion, because they are the most used invertible transforms.
The coupling blocks contain smaller feed-forward subnetworks predicting the affine coefficients. The in- and output shapes of the subnetworks depend on the in- output sizes of the coupling block itself. These sizes are not known when coding the INN (or perhaps can be worked out by hand, but would have to be worked out anew every time the architecture is modified slightly). Therefore, the subnetworks can not be directly passed as
nn.Modules, but rather in the form of a function or class, that constructs the subnetworks given in- and output size. This is a lot simpler than it sounds, for a fully connected subnetwork we could use for example:def fc_constr(dims_in, dims_out): return nn.Sequential(nn.Linear(dims_in, 128), nn.ReLU(), nn.Linear(128, 128), nn.ReLU(), nn.Linear(128, dims_out))
The RNVP and GLOW coupling blocks have an additional hyperparameter
clamp. This is because, instead of the exponential functionexp(s), we useexp( 2*c/pi * atan(x))in the coupling blocks (clamp-parameterc). This leads to much more stable training and enables larger learning rates. Effectively, the multiplication component of the coupling block is limited betweenexp(c)and1/exp(c). The Jacobian determinant is thereby limited between±D*c(dimensionality of dataD). In general,clamp = 2.0is a good place to start:glow = Node([in1.out0], GLOWCouplingBlock, {'subnet_constructor': fc_constr, 'clamp': 2.0}, name='GLOW coupling block')
Using these rules, we would construct the INN from the above example in the following way:
in1 = Ff.InputNode(100, name='Input 1') # 1D vector
in2 = Ff.InputNode(20, name='Input 2') # 1D vector
cond = Ff.ConditionNode(42, name='Condition')
def subnet(dims_in, dims_out):
return nn.Sequential(nn.Linear(dims_in, 256), nn.ReLU(),
nn.Linear(256, dims_out))
perm = Ff.Node(in1, Fm.PermuteRandom, {}, name='Permutation')
split1 = Ff.Node(perm, Fm.Split, {}, name='Split 1')
split2 = Ff.Node(split1.out1, Fm.Split, {}, name='Split 2')
actnorm = Ff.Node(split2.out1, Fm.ActNorm, {}, name='ActNorm')
concat1 = Ff.Node([actnorm.out0, in2.out0], Fm.Concat, {}, name='Concat 1')
affine = Ff.Node(concat1, Fm.AffineCouplingOneSided, {'subnet_constructor': subnet},
conditions=cond, name='Affine Coupling')
concat2 = Ff.Node([split2.out0, affine.out0], Fm.Concat, {}, name='Concat 2')
output1 = Ff.OutputNode(split1.out0, name='Output 1')
output2 = Ff.OutputNode(concat2, name='Output 2')
example_INN = Ff.GraphINN([in1, in2, cond,
perm, split1, split2,
actnorm, concat1, affine, concat2,
output1, output2])
# dummy inputs:
x1, x2, c = torch.randn(1, 100), torch.randn(1, 20), torch.randn(1, 42)
# compute the outputs
(z1, z2), log_jac_det = example_INN([x1, x2], c=c)
# invert the network and check if we get the original inputs back:
(x1_inv, x2_inv), log_jac_det_inv = example_INN([z1, z2], c=c, rev=True)
assert (torch.max(torch.abs(x1_inv - x1)) < 1e-5
and torch.max(torch.abs(x2_inv - x2)) < 1e-5)If you want full examples with training code etc., look through the experiments folder. The following only provides examples for constructing INNs by themselves.
# These imports and declarations apply to all examples
import torch.nn as nn
import FrEIA.framework as Ff
import FrEIA.modules as Fm
def subnet_fc(c_in, c_out):
return nn.Sequential(nn.Linear(c_in, 512), nn.ReLU(),
nn.Linear(512, c_out))
def subnet_conv(c_in, c_out):
return nn.Sequential(nn.Conv2d(c_in, 256, 3, padding=1), nn.ReLU(),
nn.Conv2d(256, c_out, 3, padding=1))
def subnet_conv_1x1(c_in, c_out):
return nn.Sequential(nn.Conv2d(c_in, 256, 1), nn.ReLU(),
nn.Conv2d(256, c_out, 1))The following INN only has 2 input dimensions.
It should be able to learn to generate most 2D distributions (gaussian mixtures, different shapes, ...),
and can be easily visualized.
We will use a series of AllInOneBlock operations, which combine affine coupling, a permutation and ActNorm in a single structure.
Since the computation graph is a simple chain of operations, we can define the network using the SequenceINN API.
inn = Ff.SequenceINN(2)
for k in range(8):
inn.append(Fm.AllInOneBlock, subnet_constructor=subnet_fc, permute_soft=True)The following cINN is able to perform conditional MNIST generation quite well.
Note that it is not particularly efficient, with respect to the number of parameters (see convolutional INN for that).
Again, we use a chain of AllInOneBlock``s, collected together by ``SequenceINN.
cinn = Ff.SequenceINN(28*28)
for k in range(12):
cinn.append(Fm.AllInOneBlock, cond=0, cond_shape=(10,), subnet_constructor=subnet_fc)For the following architecture (which works e.g. for CIFAR10), 3/4 of the
outputs are split off after some convolutions, which encode the local details,
and the rest are transformed further to encode semantic content. This is
important, because even for moderately sized images, it becomes infeasible to
transform all dimenions through the full depth of the INN. Many dimensions will
just enocde image noise, so we can split them off early.
Because the computational graph contains multiple outputs, we have to use the full G machinery.
nodes = [Ff.InputNode(3, 32, 32, name='input')]
ndim_x = 3 * 32 * 32
# Higher resolution convolutional part
for k in range(4):
nodes.append(Ff.Node(nodes[-1],
Fm.GLOWCouplingBlock,
{'subnet_constructor':subnet_conv, 'clamp':1.2},
name=F'conv_high_res_{k}'))
nodes.append(Ff.Node(nodes[-1],
Fm.PermuteRandom,
{'seed':k},
name=F'permute_high_res_{k}'))
nodes.append(Ff.Node(nodes[-1], Fm.IRevNetDownsampling, {}))
# Lower resolution convolutional part
for k in range(12):
if k%2 == 0:
subnet = subnet_conv_1x1
else:
subnet = subnet_conv
nodes.append(Ff.Node(nodes[-1],
Fm.GLOWCouplingBlock,
{'subnet_constructor':subnet, 'clamp':1.2},
name=F'conv_low_res_{k}'))
nodes.append(Ff.Node(nodes[-1],
Fm.PermuteRandom,
{'seed':k},
name=F'permute_low_res_{k}'))
# Make the outputs into a vector, then split off 1/4 of the outputs for the
# fully connected part
nodes.append(Ff.Node(nodes[-1], Fm.Flatten, {}, name='flatten'))
split_node = Ff.Node(nodes[-1],
Fm.Split,
{'section_sizes':(ndim_x // 4, 3 * ndim_x // 4), 'dim':0},
name='split')
nodes.append(split_node)
# Fully connected part
for k in range(12):
nodes.append(Ff.Node(nodes[-1],
Fm.GLOWCouplingBlock,
{'subnet_constructor':subnet_fc, 'clamp':2.0},
name=F'fully_connected_{k}'))
nodes.append(Ff.Node(nodes[-1],
Fm.PermuteRandom,
{'seed':k},
name=F'permute_{k}'))
# Concatenate the fully connected part and the skip connection to get a single output
nodes.append(Ff.Node([nodes[-1].out0, split_node.out1],
Fm.Concat1d, {'dim':0}, name='concat'))
nodes.append(Ff.OutputNode(nodes[-1], name='output'))
conv_inn = Ff.GraphINN(nodes)Custom invertible modules can be written as extensions of the Fm.InvertibleModule base class. Refer to the documentation of this class for detailed information on requirements.
Below are two simple examples which illustrate the definition and use of custom modules and can be used as basic templates. The first multiplies each dimension of an input tensor by either 1 or 2, chosen in a random but fixed way. The second is a conditional operation which takes two inputs and swaps them if the condition is positive, doing nothing otherwise.
Notes:
- The
Fm.InvertibleModulemust be initialized with thedims_inargument and optionallydims_cif there is a conditioning input. forwardshould return a tuple of outputs (even if there is only one), with additionallog_jac_detterm. This Jacobian term can be, but does not need to be calculated ifjac=False.
Definition:
class FixedRandomElementwiseMultiply(Fm.InvertibleModule):
def __init__(self, dims_in):
super().__init__(dims_in)
self.random_factor = torch.randint(1, 3, size=(1, dims_in[0][0]))
def forward(self, x, rev=False, jac=True):
# the Jacobian term is trivial to calculate so we return it
# even if jac=False
# x is passed to the function as a list (in this case of only on element)
x = x[0]
if not rev:
# forward operation
x = x * self.random_factor
log_jac_det = self.random_factor.float().log().sum()
else:
# backward operation
x = x / self.random_factor
log_jac_det = -self.random_factor.float().log().sum()
return (x,), log_jac_det
def output_dims(self, input_dims):
return input_dims
class ConditionalSwap(Fm.InvertibleModule):
def __init__(self, dims_in, dims_c):
super().__init__(dims_in, dims_c=dims_c)
def forward(self, x, c, rev=False, jac=True):
# in this case, the forward and reverse operations are identical
# so we don't use the rev argument
x1, x2 = x
log_jac_det = 0.
# make copies of the inputs
x1_new = x1 + 0.
x2_new = x2 + 0.
for i in range(x1.size(0)):
x1_new[i] = x1[i] if c[0][i] > 0 else x2[i]
x2_new[i] = x2[i] if c[0][i] > 0 else x1[i]
return (x1_new, x2_new), log_jac_det
def output_dims(self, input_dims):
return input_dimsBasic Usage Example:
BATCHSIZE = 10
DIMS_IN = 2
# build up basic net using SequenceINN
net = Ff.SequenceINN(DIMS_IN)
for i in range(2):
net.append(FixedRandomElementwiseMultiply)
# define inputs
x = torch.randn(BATCHSIZE, DIMS_IN)
# run forward
z, log_jac_det = net(x)
# run in reverse
x_rev, log_jac_det_rev = net(z, rev=True)More Complicated Example:
BATCHSIZE = 10
DIMS_IN = 2
# define a graph INN
input_1 = Ff.InputNode(DIMS_IN, name='input_1')
input_2 = Ff.InputNode(DIMS_IN, name='input_2')
cond = Ff.ConditionNode(1, name='condition')
mult_1 = Ff.Node(input_1.out0, FixedRandomElementwiseMultiply, {}, name='mult_1')
cond_swap = Ff.Node([mult_1.out0, input_2.out0], ConditionalSwap, {}, conditions=cond, name='conditional_swap')
mult_2 = Ff.Node(cond_swap.out1, FixedRandomElementwiseMultiply, {}, name='mult_2')
output_1 = Ff.OutputNode(cond_swap.out0, name='output_1')
output_2 = Ff.OutputNode(mult_2.out0, name='output_2')
net = Ff.GraphINN([input_1, input_2, cond, mult_1, cond_swap, mult_2, output_1, output_2])
# define inputs
x1 = torch.randn(BATCHSIZE, DIMS_IN)
x2 = torch.randn(BATCHSIZE, DIMS_IN)
c = torch.randn(BATCHSIZE)
# run forward
(z1, z2), log_jac_det = net([x1, x2], c=c)
# run in reverse without necessarily calculating Jacobian term (i.e. jac=False)
(x1_rev, x2_rev), _ = net([z1, z2], c=c, rev=True, jac=False)- Stochastic gradient descent will not work (well) for INNs. Use e.g. Adam instead.
- Gradient clipping can be useful if you are experiencing training instabilities, e.g. use
torch.nn.utils.clip_grad_norm_. - Add some slight noise to the inputs (order of 1E-2). This stabilizes training and prevents sparse gradients, if there are some quantized or perfectly correlated input dimensions.
For coupling blocks in particular:
- Use Xavier initialization for the weights. This prevents unstable training at the start.
- If your network is very deep (>30 coupling blocks), initialize the last layer in the subnetworks to zero. This means the INN as a whole is initialized to the identity, and you will not get NaNs at the first iteration.
- Do not forget permutations/orthogonal transforms between coupling blocks.
- Keep the subnetworks shallow (2-3 layers only), but wide (>= 128 neurons/ >= 64 conv. channels).
- Keep in mind that one coupling block contains between 4 and 12 individual convolutions or fully connected layers. So you may not have to use as many as you think, else the number of parameters will be huge.
- This being said, as the coupling blocks initialize to roughly the identity transform, it is hard to have too many coupling blocks and break the training completely (as opposed to a standard feed-forward NN).
For convolutional INNs in particular:
- Perform some kind of reshaping early, so the INN has >3 channels to work with.
- Coupling blocks using 1x1 convolutions in the subnets seem important for the quality, they should constitute every other, or every third coupling block.
