This repository is the official NumPy implementation of the paper Deep Networks from the Principle of Rate Reduction (2021) by Kwan Ho Ryan Chan* (UC Berkeley), Yaodong Yu* (UC Berkeley), Chong You* (UC Berkeley), Haozhi Qi (UC Berkeley), John Wright (Columbia), and Yi Ma (UC Berkeley). For PyTorch version of ReduNet, please visit https://github.com/ryanchankh/redunet.
ReduNet is a deep neural network construcuted naturally by deriving the gradients of the Maximal Coding Rate Reduction (MCR2) [1] objective. Every layer of this network can be interpreted based on its mathematical operations and the network collectively is trained in a feed-forward manner only. In addition, by imposing shift invariant properties to our network, the convolutional operator can be derived using only the data and MCR2 objective function, hence making our network design principled and interpretable.
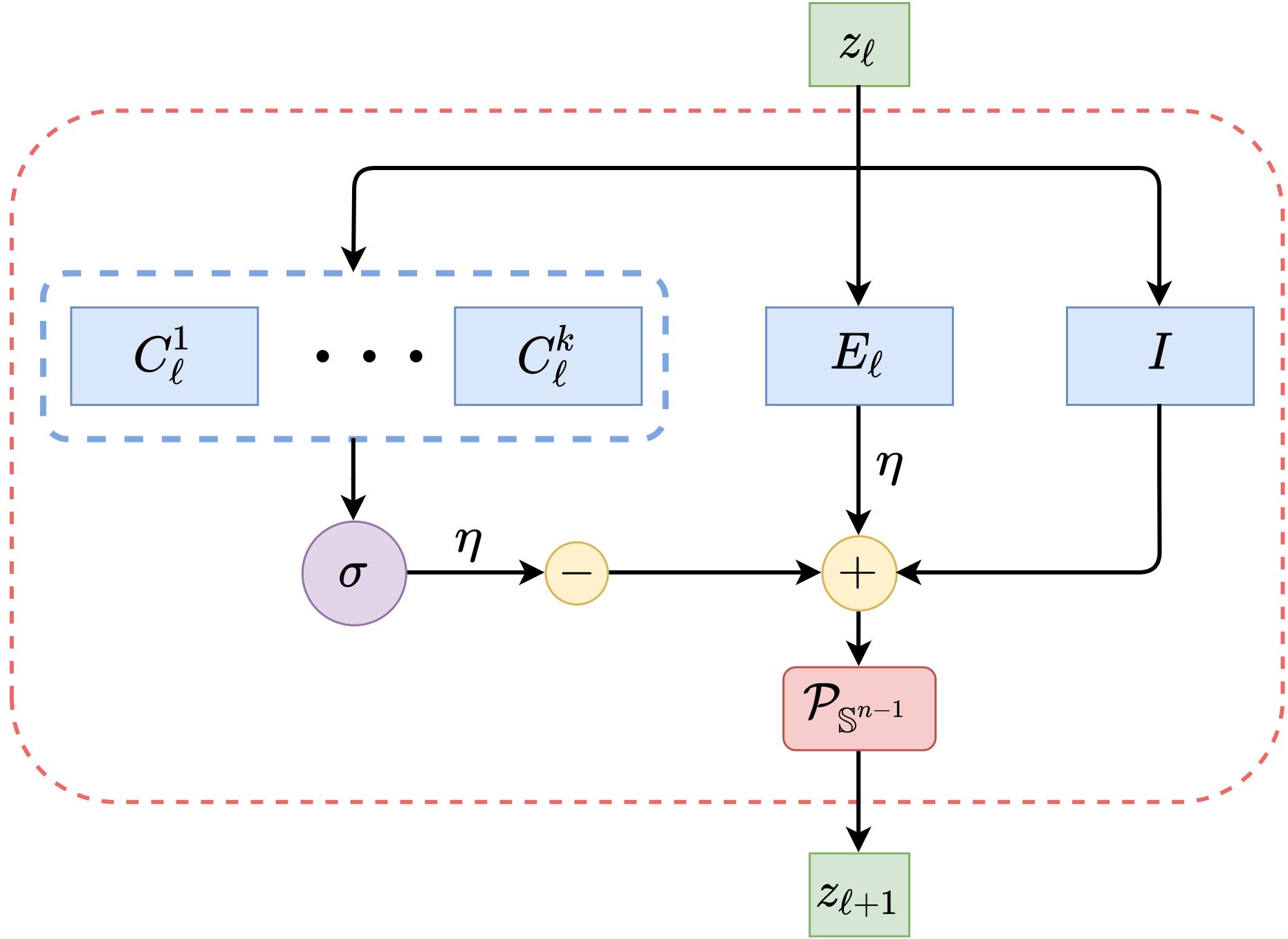
Figure: Weights and operations for one layer of ReduNet
[1] Yu, Yaodong, Kwan Ho Ryan Chan, Chong You, Chaobing Song, and Yi Ma. "Learning diverse and discriminative representations via the principle of maximal coding rate reduction" Advances in Neural Information Processing Systems 33 (2020).
This codebase is written for python3. To install necessary python packages, run conda create --name redunet_official --file requirements.txt.
To train a model, one can run the training files, which has the dataset as thier names. For the appropriate commands to reproduce our experimental results, check out the experiment section below. All the files for training is listed below:
gaussian2d.py: mixture of Guassians in 2-dimensional Realsgaussian3d.py: mixture of Guassians in 3-dimensional Realsiris.py: Iris dataset from UCI Machine Learning Repository (link)mice.py: Mice Protein Expression Data Set (link)mnist1d.py: MNIST dataset, each image is multi-channel polar form and model is trained to have rotational invariancemnist2d.py: MNIST dataset, each image is single-channel and model is trained to have translational invariancesinusoid.py: mixture of sinusoidal waves, single and multichannel data
Evaluation and plots are performed within each file. Functions are located in evaluate.py and plot.py.
Run the following commands to train, test, evaluate and plot figures for different settings:
Gaussian 2D: Figure 2(a) - (c)
$ python3 gaussian2d.py --data 1 --noise 0.1 --samples 500 --layers 2000 --eta 0.5 --eps 0.1
Gaussian 3D: Figure 2(d) - (f)
$ python3 gaussian3d.py --data 1 --noise 0.1 --samples 500 --layers 2000 --eta 0.5 --eps 0.1
Rotational-Invariant MNIST: 3(a) - (d)
$ python3 mnist1d.py --samples 10 --channels 15 --outchannels 20 --time 200 --classes 0 1 2 3 4 5 6 7 8 9 --layers 40 --eta 0.5 --eps 0.1 --ksize 5
Translational-Invariant MNIST: 3(e) - (h)
$ python3 mnist2d.py --classes 0 1 2 3 4 5 6 7 8 9 --samples 10 --layers 25 --outchannels 75 --ksize 9 --eps 0.1 --eta 0.5
For Iris and Mice Protein:
$ python3 iris.py --layers 4000 --eta 0.1 --eps 0.1
$ python3 mice.py --layers 4000 --eta 0.1 --eps 0.1
For 1D signals (Sinusoids):
$ python3 sinusoid.py --time 150 --samples 400 --channels 7 --layers 2000 --eps 0.1 --eta 0.1 --data 7 --kernel 3
For 1D signals (Rotational Invariant MNIST):
$ python3 mnist1d.py --classes 0 1 --samples 2000 --time 200 --channels 5 --layers 3500 --eta 0.5 --eps 0.1
For 2D translational invariant MNIST data:
$ python3 mnist2d.py --classes 0 1 --samples 500 --layers 2000 --eta 0.5 --eps 0.1
For technical details and full experimental results, please check the paper. Please consider citing our work if you find it helpful to yours:
@article{chan2020deep,
title={Deep networks from the principle of rate reduction},
author={Chan, Kwan Ho Ryan and Yu, Yaodong and You, Chong and Qi, Haozhi and Wright, John and Ma, Yi},
journal={arXiv preprint arXiv:2010.14765},
year={2020}
}
- This README is formatted based on paperswithcode.
- Feel free to post issues via Github.
Please contact ryanchankh@berkeley.edu and yyu@eecs.berkeley.edu if you have any question on the codes.