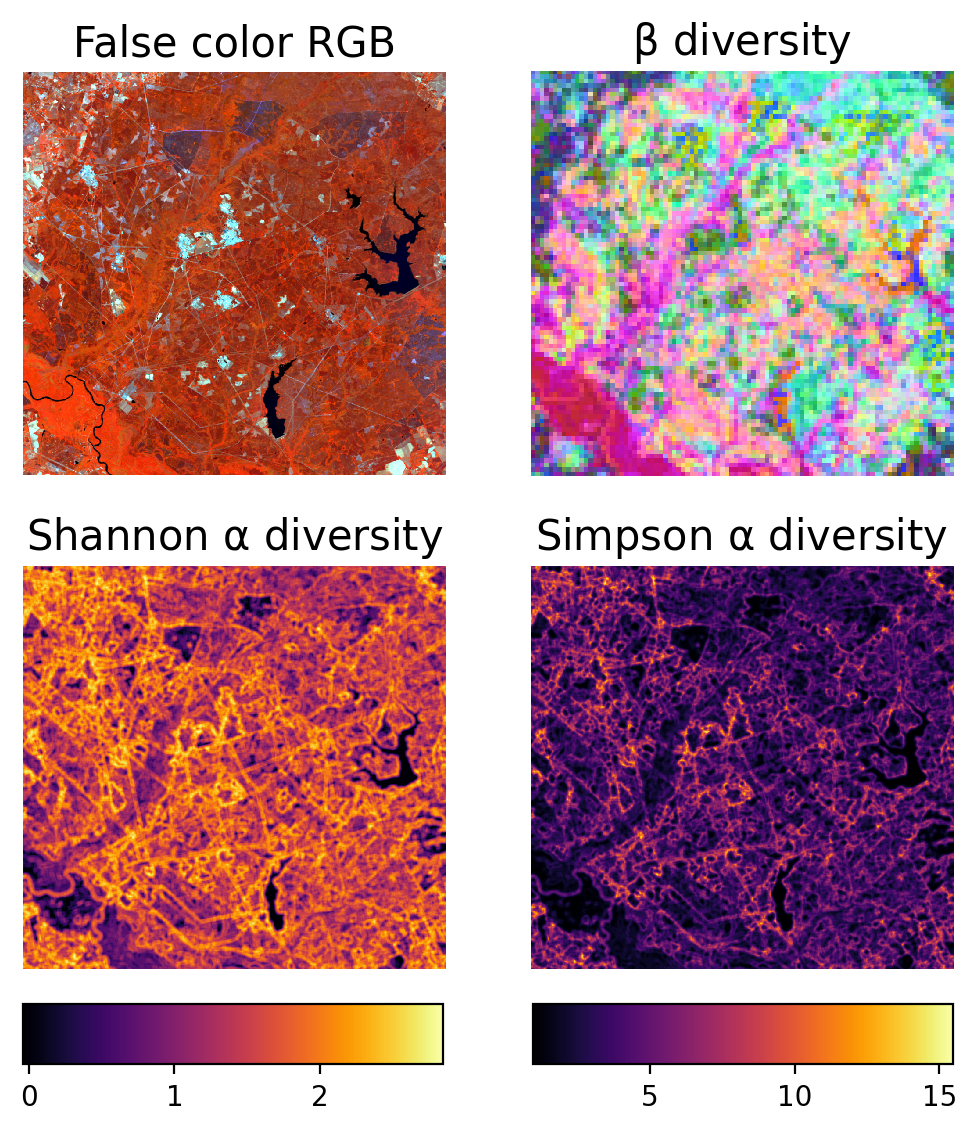
sister-biodiversity
This repository contains code for calculating alpha and beta spectral diversity metrics. Methods were adapted from Féret & de Boissieu (2020) but do not exactly replicate their techniques. We have included a command line script, which can automatically generate diversity metrics with user specfied parameters, along with an interactive Jupyter notebook (recommended).
Féret, J. B., & de Boissieu, F. (2020). biodivMapR: An r package for α‐and β‐diversity mapping using remotely sensed images. Methods in Ecology and Evolution, 11(1), 64-70. https://doi.org/10.1111/2041-210X.13310
Installation
pip -r requirements.txtUse
python spectral_diversity.py reflectance_image output_directoryOptional arguments:
--nclusters: Number of k-means clusters, default = 25--window: Window size for calculating diversity metrics, default= 10--pca: Export PCA image, default = False--species: Export spectral species map, default = False--ncpus: Number of CPUs for MDS, default = 1--verbose: default = False