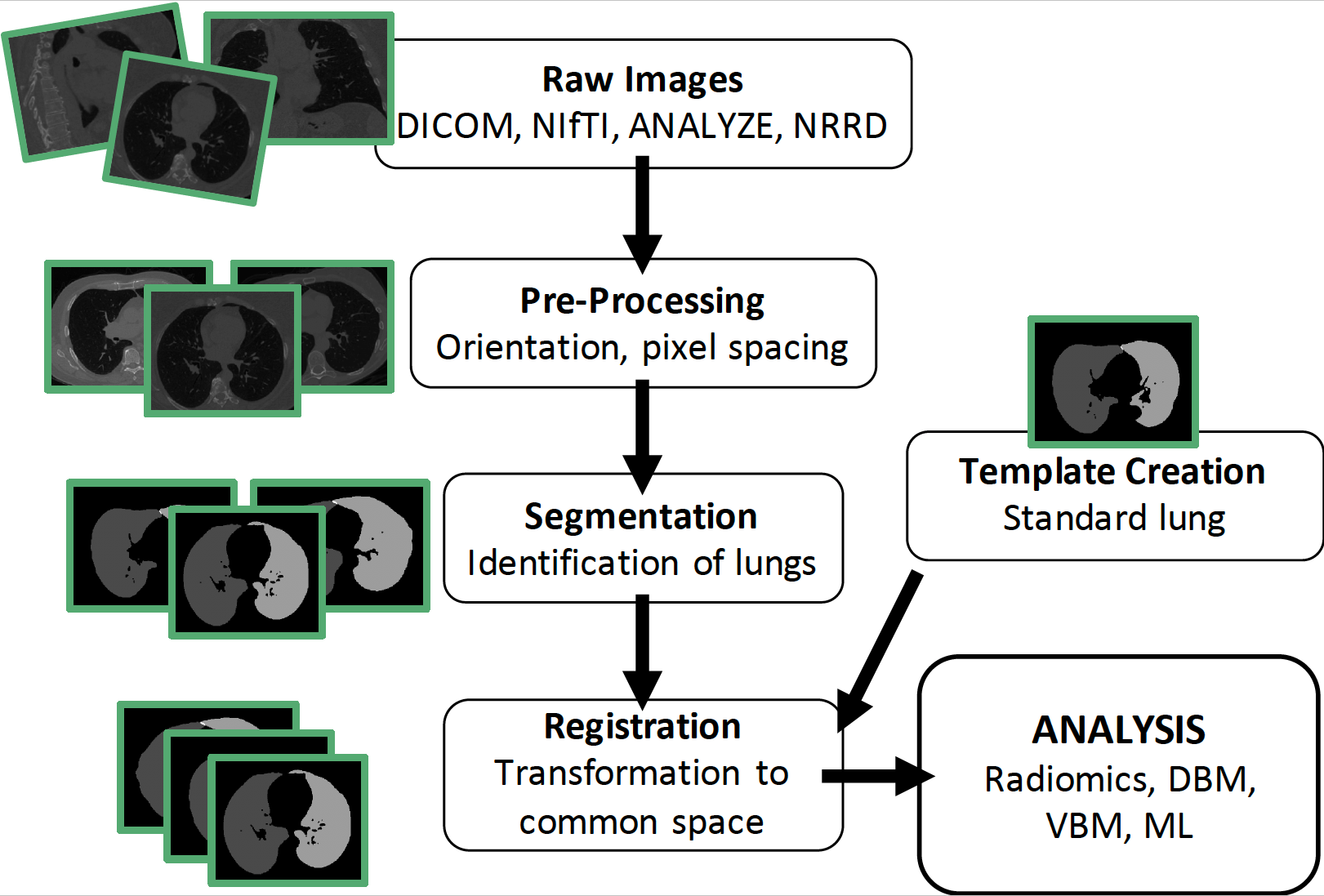
The lungct R package develops an image processing pipeline for computed tomography (CT) scans of the lungs.
-
We develop a free and simple segmentation algorithm that is comparable to the proprietary VIDA Diagnostics software
-
We create the first publicly available standard lung template using healthy adults, which is available for download via lungct
-
We show that the standard lung template allows for improved population-level inference of lung CTs using radiomics
-
lungct provides a fully-automated and open-source image processing pipeline for lung CTs, which is accessible to statisticians
You can install lungct from github with:
# install.packages("devtools")
devtools::install_github("muschellij2/lungct")To segment the lungs from the CT scan:
library(lungct)
filename <- "example.nii.gz"
mask <- segment_lung(filename)To segment the left and right lungs from the CT scan:
img <- ANTsRCore::antsImageRead(filename)
mask2 <- segment_lung2(img)The standard lung was created from N=62 healthy controls from COPDGene (50% female, 95% white, mean age = 62 years, mean BMI = 28.5)
To load the standard lung:
# Read in standard lung template
filepath <- system.file(
"extdata", "lung_template_mask.nii.gz",
package = "lungct")
template <- ANTsRCore::antsImageRead(filepath)To register the mask to the standard lung:
# Register mask to standard lung
reg <- register_lung_mask(
moving_mask = mask,
fixed_mask = template,
moving = img,
sides = c("right","left"),
typeofTransform = "SyN")We recommend the following for lung registration:
-
Separately register the right and left lungs to account for differences in lung shape and size
-
Perform registration on lung masks to preserve biological variability in Hounsfield units (HU)
-
Use Symmetric Normalization (SyN) for nonlinear registration due to its flexibility and success in the EMPIRE10 challenge
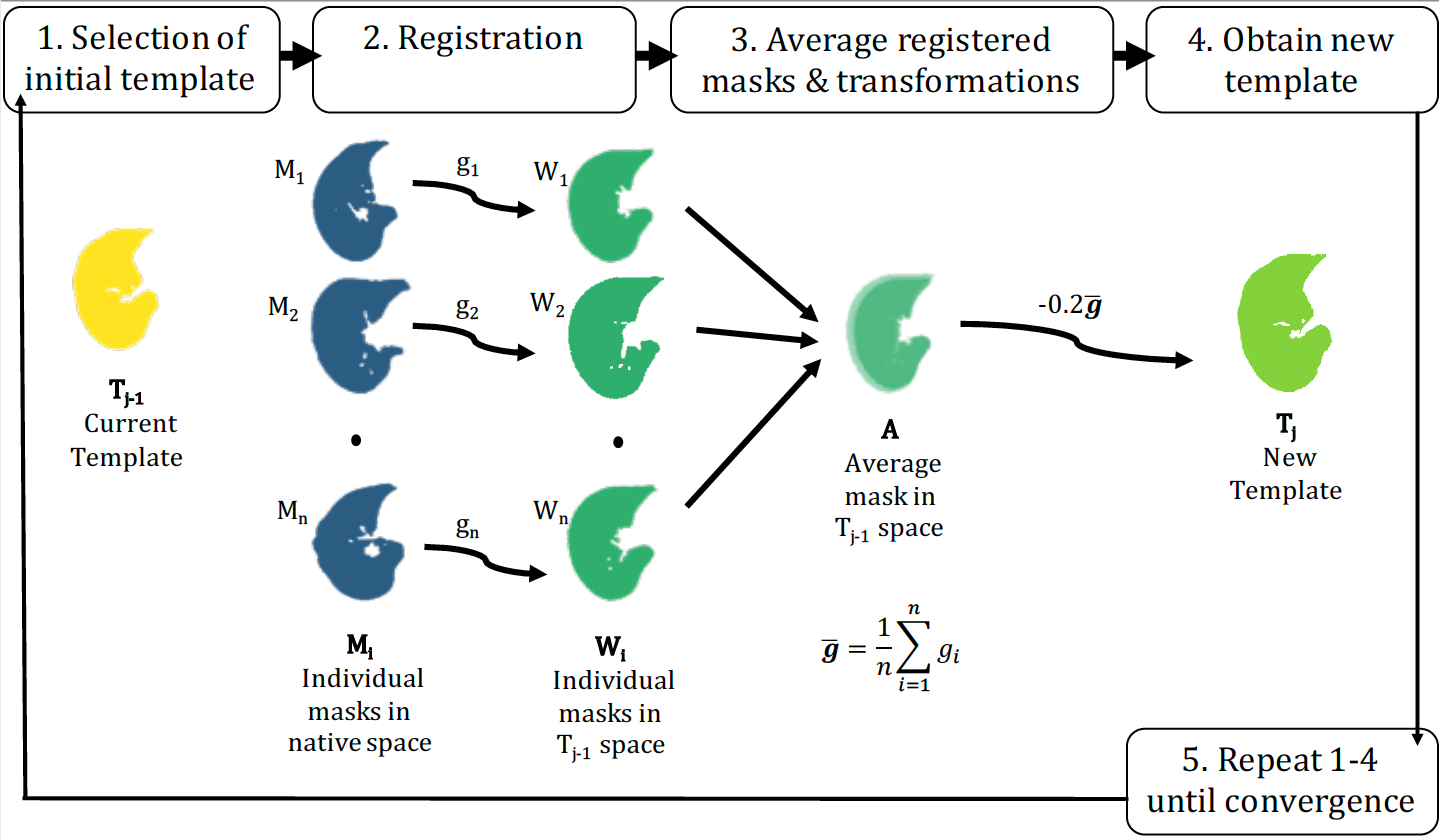
For template creation, we follow the iterative method from Avants, using iterations of register_lung_mask, get_template, and calculate_DSC from lungct. However, we define convergence as having a Dice similarity coefficient (DSC) between successive iterations of at least 0.99. We recommend using parallelization for the registration step.
# Obtain new template
template_new <- get_template(
folder_warp = warped_masks,
folder_comp = transformations)
# Check convergence
dice <- calculate_DSC(template_init, template_new)Radiomics, a field of quantitative imaging where large amounts of features are extracted from medical images, is common for lung CTs.
Options in lungct:
-
radiomics_slice: Calculation on 2D slices in axial, coronal, or sagittal planes
-
radiomics_lung: Calculation on 3D right and left lungs
-
RIA_lung: Calculation of advanced radiomic features, such as GLCM and GLRLM More info
# Calculate radiomics
rad <- RIA_lung(
img, mask,
sides = c("right", "left"),
features = c("fo", "glcm"),
bins_in = 16, equal_prob = FALSE, distance = 1,
statistic = "mean(X, na.rm = TRUE)")For the lung CT template, the COPDGene data was used:
"This research used data generated by the COPDGene study, which was supported by NIH grants U01HL089856 and U01HL089897. The COPDGene project is also supported by the COPD Foundation through contributions made by an Industry Advisory Board comprised of Pfizer, AstraZeneca, Boehringer Ingelheim, Novartis, and Sunovion."