Authors: Bruno Aristimunha, Alexandre Bayerlein, M. Jorge Cardoso, Walter Lopez Pinaya, Raphael Yokoingawa de Camargo.
IEEE Access, Accepted March, 2023.
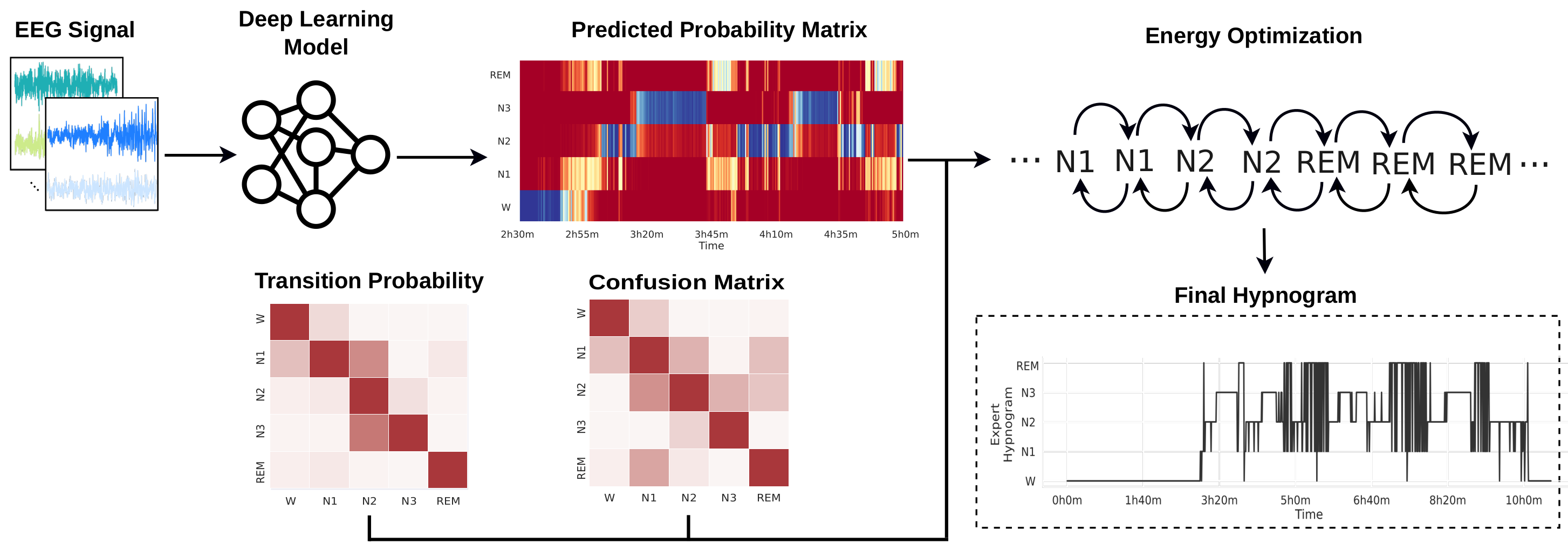
Sleep is essential for physical and mental health. Polysomnography (PSG) procedures are labour-intensive and time-consuming, making diagnosing sleep disorders difficult. Automatic sleep staging using ML-based methods has been studied extensively, but frequently provides noisier predictions incompatible with typical manually annotated hypnograms. We propose an energy optimization method to improve the quality of hypnograms generated by automatic sleep staging procedures. The method evaluates the system’s total energy based on conditional probabilities for each epoch’s stage and employs an energy minimisation procedure. It can be used as a meta-optimisation layer over the sleep stage sequences generated by any classifier that generates prediction probabilities. Results show that it improves the accuracy of the predictions of state-of-the-art Deep Learning models using two public datasets.
To execute the model, you can use the script src/energy-model.py.
Example: energy-model.py --writefile 1 --optparams 0 --model usleep --part 0 --dataset edf
Parameters:
'--optparams', type=int, default=0,
Set 1 to run the energy model with multiple alpha values and 0 for a single one (alpha = 0.5).
'--part', type=int, default=0,
Define the CV slice to optimize, between 0 and 5. Use -1 for all parts.
'--model', type=str, default='usleep',
Name of the neural network model ['stager', 'usleep', 'all']
'--writefile', type=int, default=1,
Set 1 to write results to files and 0 otherwise.
'--dataset', type=str, default='edf',
Name of the dataset to optimize ['edf', 'dreamer', 'all']
Input files: Located in data folder
input_[dataset]_[model]_part_[0-4].pkl
A pickle file containing a DataFrame data_per_subject with 5 columns and one line per subject and sleep session:
- subjects: a pair (subject, session) identifying the subject and session
- y_true: The hypnogram gerated by an specialist (target hypnogram)
- y_pred: The predicted hypnogram using the neural network
model - y_prob: The probabilities generated by
modelfor each possible stage in each epoch - train_test: Indicates if the hypnogram was used as part of the
train,valid, ortestsets
There is one file per model and dataset. Also, we used 5-fold cross-validation, and each part represents one fold.
Output files: Generated at output folder
sleep-results-[dataset].csv: A csv file with multiple metrics, such as accuracy, f1-score,
precision, recall, and balanced accuracy. Each line contains the results for a single subject and sleep record,
in addition to parameter values and neural network model used for the predictions.
energy-[model]-[0-4]-[dataset]-opt.npz: Contains numpy matrices with information from the energy-optimization
process, including the confusion and transition probability matrices, the target hypnograms,
the hypnograms predicted by the NN, and the hypnograms optimized byt the energy model.