Dna Features Viewer is a Python library to (wait for it...) visualize DNA features, e.g. from GenBank or Gff files, using the plotting library Matplotlib:
Dna Features Viewer is meant to automatically produce simple and clear plots even for sequences with lots of overlapping features and long labels. The plots can be output to many different formats (PNG, JPEG, SVG, PDF), e.g. for report generation or LIMS interfaces.
Dna Features Viewer is an open-source software originally written at the Edinburgh Genome Foundry by Zulko and released on Github under the MIT licence. Everyone is welcome to contribute !
If you have PIP installed, just type in a terminal:
(sudo) pip install dna_features_viewerDna Features Viewer can be installed by unzipping the source code in one directory and using this command:
sudo python setup.py installIn this first example we define features "by hand":
from dna_features_viewer import GraphicFeature, GraphicRecord
features=[
GraphicFeature(start=0, end=20, strand=+1, color="#ffd700",
label="Small feature"),
GraphicFeature(start=20, end=500, strand=+1, color="#ffcccc",
label="Gene 1 with a very long name"),
GraphicFeature(start=400, end=700, strand=-1, color="#cffccc",
label="Gene 2"),
GraphicFeature(start=600, end=900, strand=+1, color="#ccccff",
label="Gene 3")
]
record = GraphicRecord(sequence_length=1000, features=features)
record.plot(fig_width=5)If we replace GraphicRecord by CircularGraphicRecord in the code above we obtain a circular plot of the construct:
It is also possible to generate interactive (browser-based) plots by using plot_with_bokeh instead of plot:
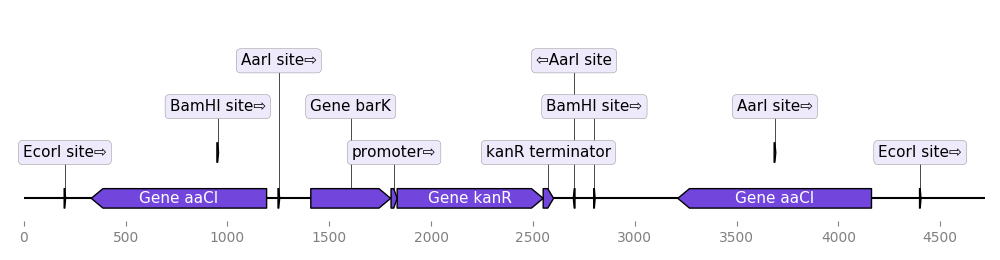
DnaFeaturesViewer plays nice with BioPython. As a result it is super easy to plot the content of a Biopython record or directly a GenBank file:
from dna_features_viewer import BiopythonTranslator
graphic_record = BiopythonTranslator().translate_record("my_sequence.gb")
ax, _ = graphic_record.plot(figure_width=10)The class BiopythonTranslator determines how the genbank information is transformed into graphical features. It enables to chose which categories of features to plot, the color of the different features.
As it uses Matplotlib, Dna Features Viewer can display the features on top of other sequences statistics, such as the local GC content:
import matplotlib.pyplot as plt
from dna_features_viewer import BiopythonTranslator
from Bio import SeqIO
import numpy as np
fig, (ax1, ax2) = plt.subplots(2, 1, figsize=(8, 4), sharex=True)
# Parse the genbank file, plot annotations
record = SeqIO.read("example_sequence.gb", "genbank")
graphic_record = BiopythonTranslator().translate_record(record)
ax, levels = graphic_record.plot()
graphic_record.plot(ax=ax1, with_ruler=False)
# Plot the local GC content
def plot_local_gc_content(record, window_size, ax):
gc_content = lambda s: 100.0*len([c for c in s if c in "GC"]) / len(s)
yy = [gc_content(record.seq[i:i+window_size])
for i in range(len(record.seq)-window_size)]
xx = np.arange(len(record.seq)-window_size)+25
ax.fill_between(xx, yy, alpha=0.3)
ax.set_ylabel("GC(%)")
plot_local_gc_content(record, window_size=50, ax=ax2)
# Resize the figure
fig.savefig("with_plot.png")Dna Features Viewer is pretty minimal in terms of features but easily extensible since it uses Matplotlib as a backend.