Healthsea is a pipeline for analyzing user reviews to supplement products by extracting their effects on health.
Learn more about Healthsea in our blog post!
Healthsea aims to analyze user-written reviews of supplements in relation to their effects on health. Based on this analysis, we try to provide product recommendations. For many people, supplements are an addition to maintaining health and achieving personal goals. Due to their rising popularity, consumers have increasing access to a variety of products.
However, it's likely that most of the products on the market are redundant or produced in a "quantity over quality" fashion to maximize profit. The resulting white noise of products makes it hard to find the right supplements.
Healthsea automizes the analysis and provides information in a more digestible way. ✨
To run this project you need:
spacy>=3.2.0
benepar>=0.2.0
torch>=1.6.0
spacy-transformers>=1.1.2
You can install them in the project folder via spacy project run install
| Documentation | |
|---|---|
| 🧭 Usage | How to use the pipeline |
| ⚙️ Pipeline | Learn more about the architecture of the pipeline |
| 🪐 spaCy project | Introduction to the spaCy project |
| ✨ Demos | Introduction to the Healthsea demos |
The pipeline processes reviews to supplements and returns health effects for every found health aspect.
You can either train the pipeline yourself with the provided datasets in the spaCy project or directly download the trained Healthsea pipeline from Huggingface via
pip install https://huggingface.co/explosion/en_healthsea/resolve/main/en_healthsea-any-py3-none-any.whl
import spacy
nlp = spacy.load("en_healthsea")
doc = nlp("This is great for joint pain.")
# Clause Segmentation & Blinding
print(doc._.clauses)
> {"split_indices": [0, 7],
> "has_ent": true,
> "ent_indices": [4, 6],
> "blinder": "_CONDITION_",
> "ent_name": "joint pain",
> "cats": {
> "POSITIVE": 0.9824668169021606,
> "NEUTRAL": 0.017364952713251114,
> "NEGATIVE": 0.00002889777533710003,
> "ANAMNESIS": 0.0001394189748680219
> },
> "prediction_text": ["This", "is", "great", "for", "_CONDITION_", "!"]}
# Aggregated results
print(doc._.health_effects)
> {"joint_pain": {
> "effects": ["POSITIVE"],
> "effect": "POSITIVE",
> "label": "CONDITION",
> "text": "joint pain"
> }}
The pipeline consists of the following components:
pipeline = [sentencizer, tok2vec, ner, benepar, segmentation, clausecat, aggregation]
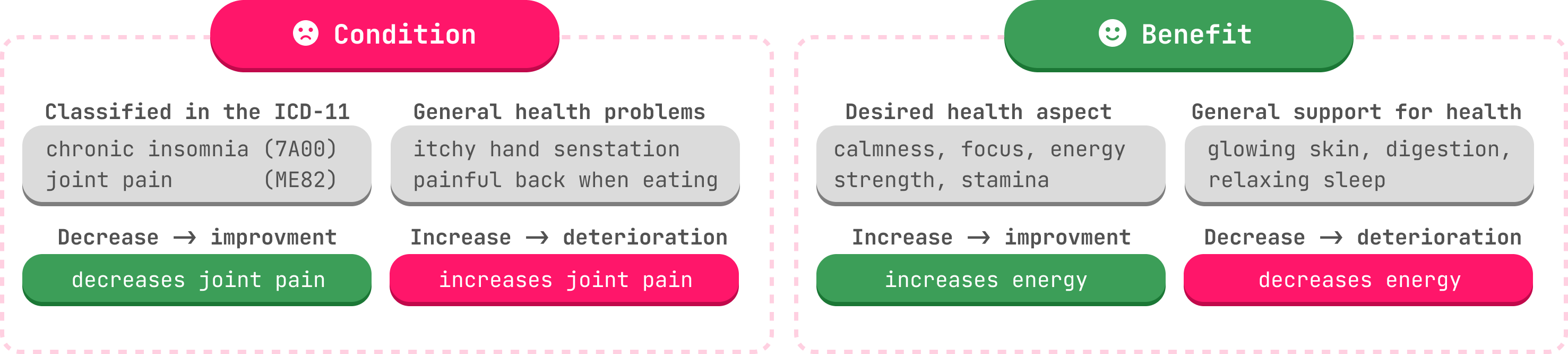
It uses Named Entity Recognition to detect two types of entities Condition and Benefit.
Condition entities are defined as health aspects that are improved by decreasing them. They include diseases, symptoms and general health problems (e.g. pain in back). Benefit entities on the other hand, are desired states of health (muscle recovery, glowing skin) that improve by increasing them.
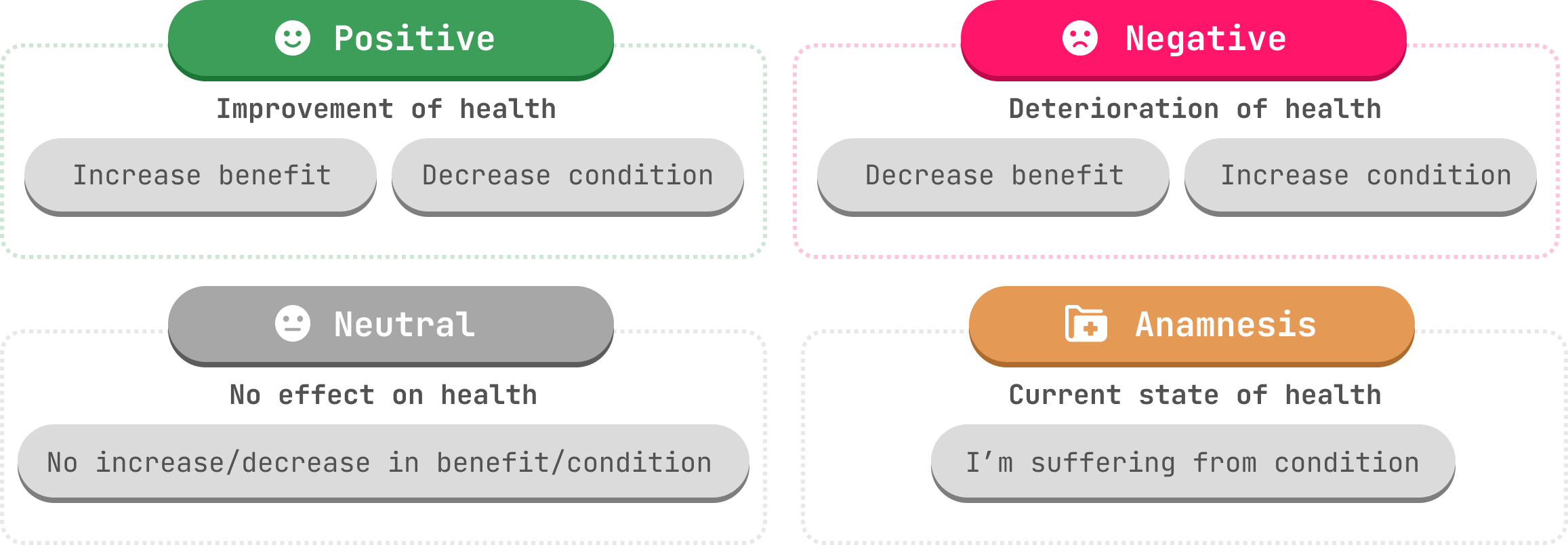
The pipeline uses a modified model that performs Clause Segmentation based on the benepar parser, Entity Blinding and Text Classification. It predicts four exclusive effects: Positive, Negative, Neutral, and Anamnesis.
The project folder contains a spaCy project with all the training data and workflows.
Use spacy project run inside the project folder to get an overview of all commands and assets. For more detailed documentation, visit the project folders readme.
Use spacy project run install to install dependencies needed for the pipeline.
A demo for exploring the results of Healthsea on real data can be found at Hugging Face Spaces.
A demo for exploring the Healthsea pipeline with its individual processing steps can be found at Hugging Face Spaces.