Bash and Python script for data-extraction from FIA-MS reports on a Agilent HPLC-MS system for HTS for unlimited number of samples (in this case 96). It is recommended to use the bash script. The python-version can be used to just extract the data from the corresponding text-files after they were extracted with the bash-script extract-data. The script evaluation converts the pdf-report to a text file, using pdftotext (a command line tool). From this text files, the corresponding mass signals are extracted and printed to a simple output file, containing the mass signals in columns. This columns can then be pasted into an excel-template for further data evaluation.
If you have questions, please feel free to message me on Github or per mail: sarah.berger@uni-graz.at
- Windows 10 or Windows 11
- Windows Subsystem Linux (WSL)
- Install Visual Studio Code (VSC)
-
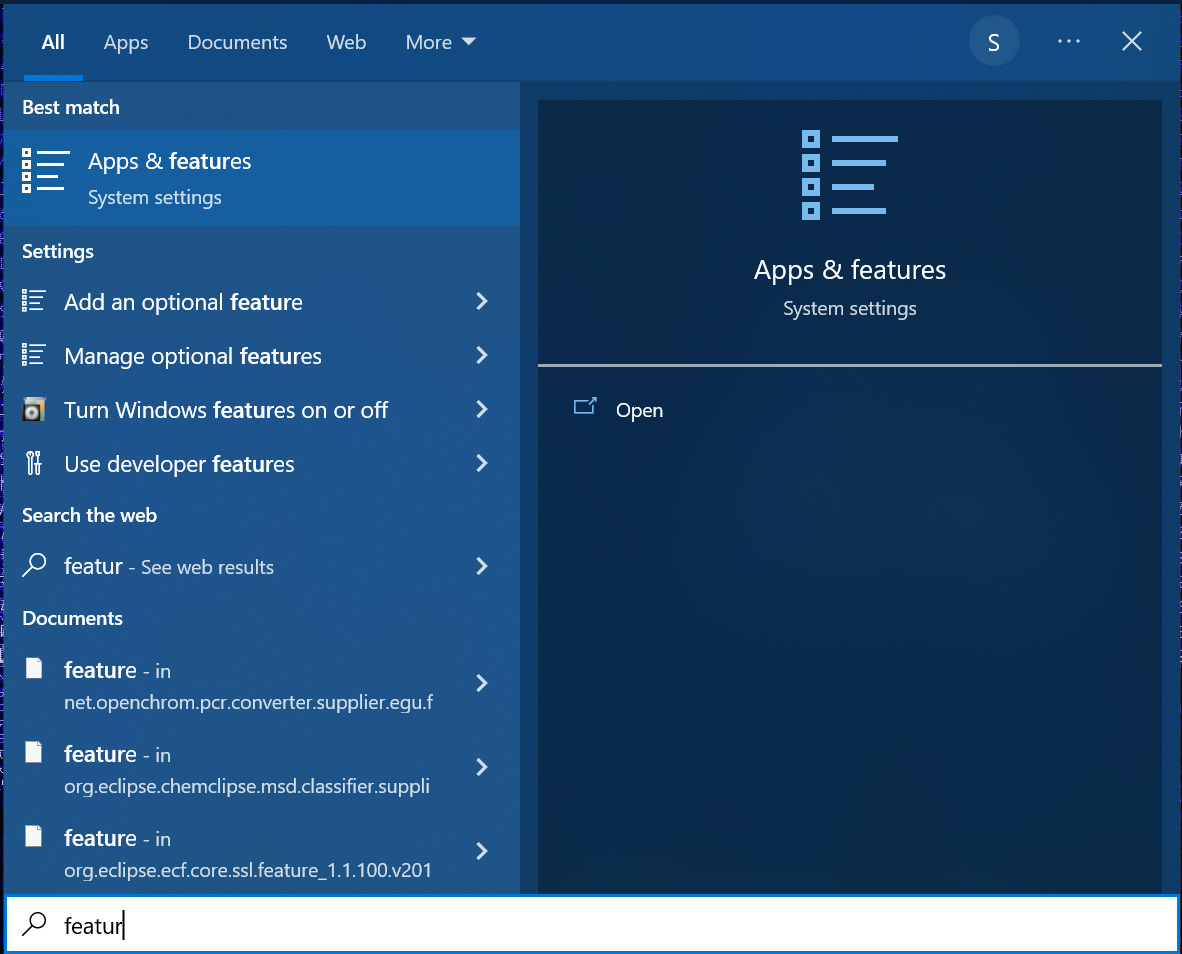
Select Turn Windows features on or off
-
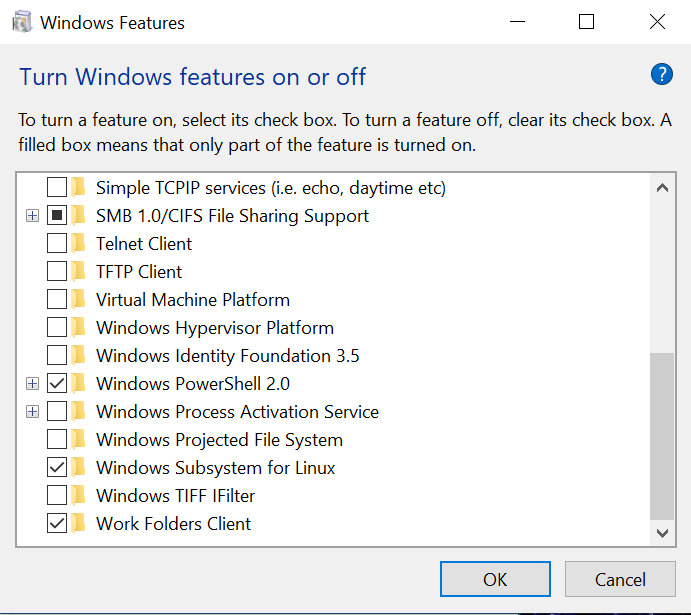
Scroll to the very bottom and check Windows Subsystem for Linux (reboot required)
-
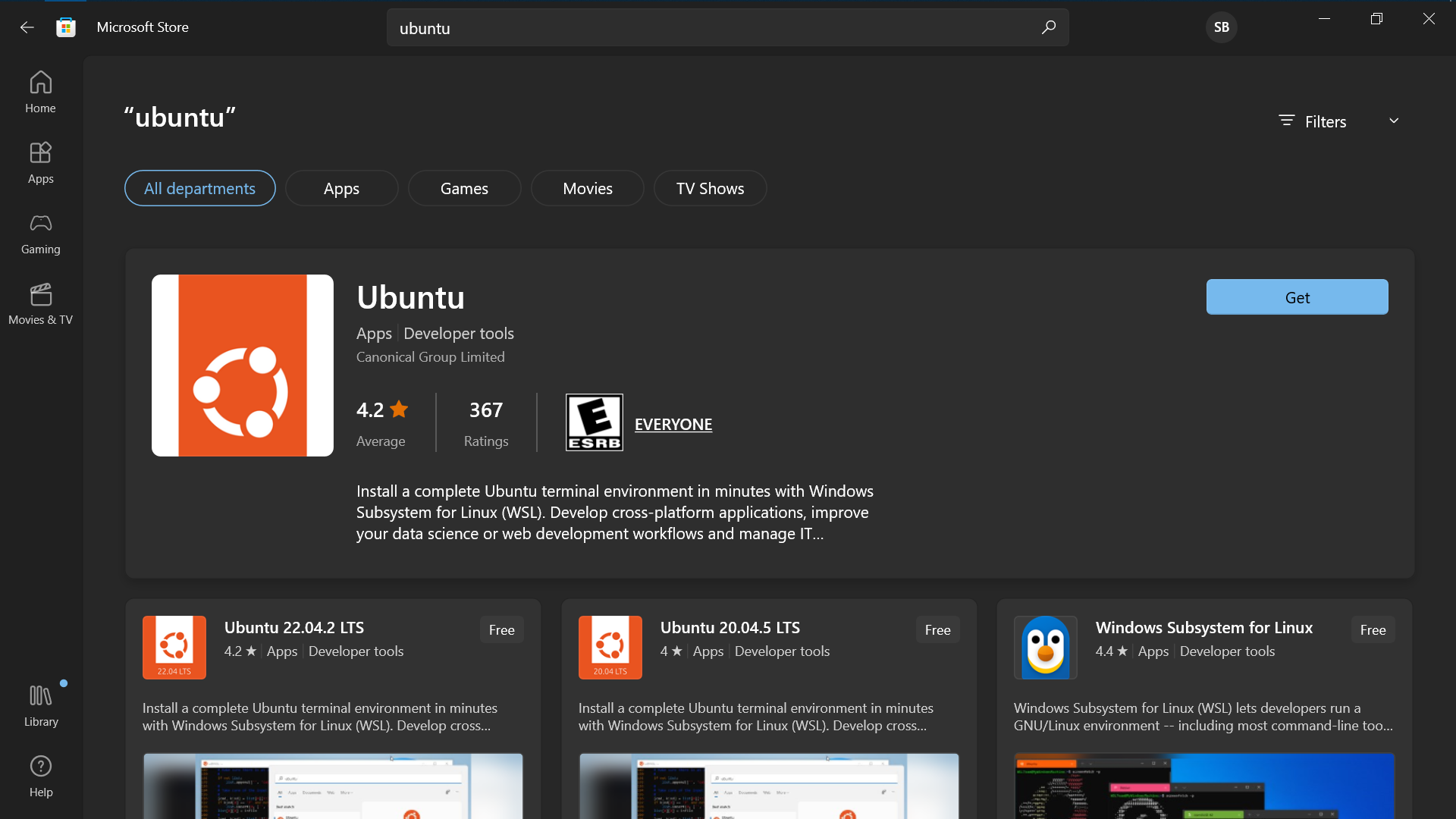
Install Ubuntu from the Microsoft Store (version tested 18.04.5)
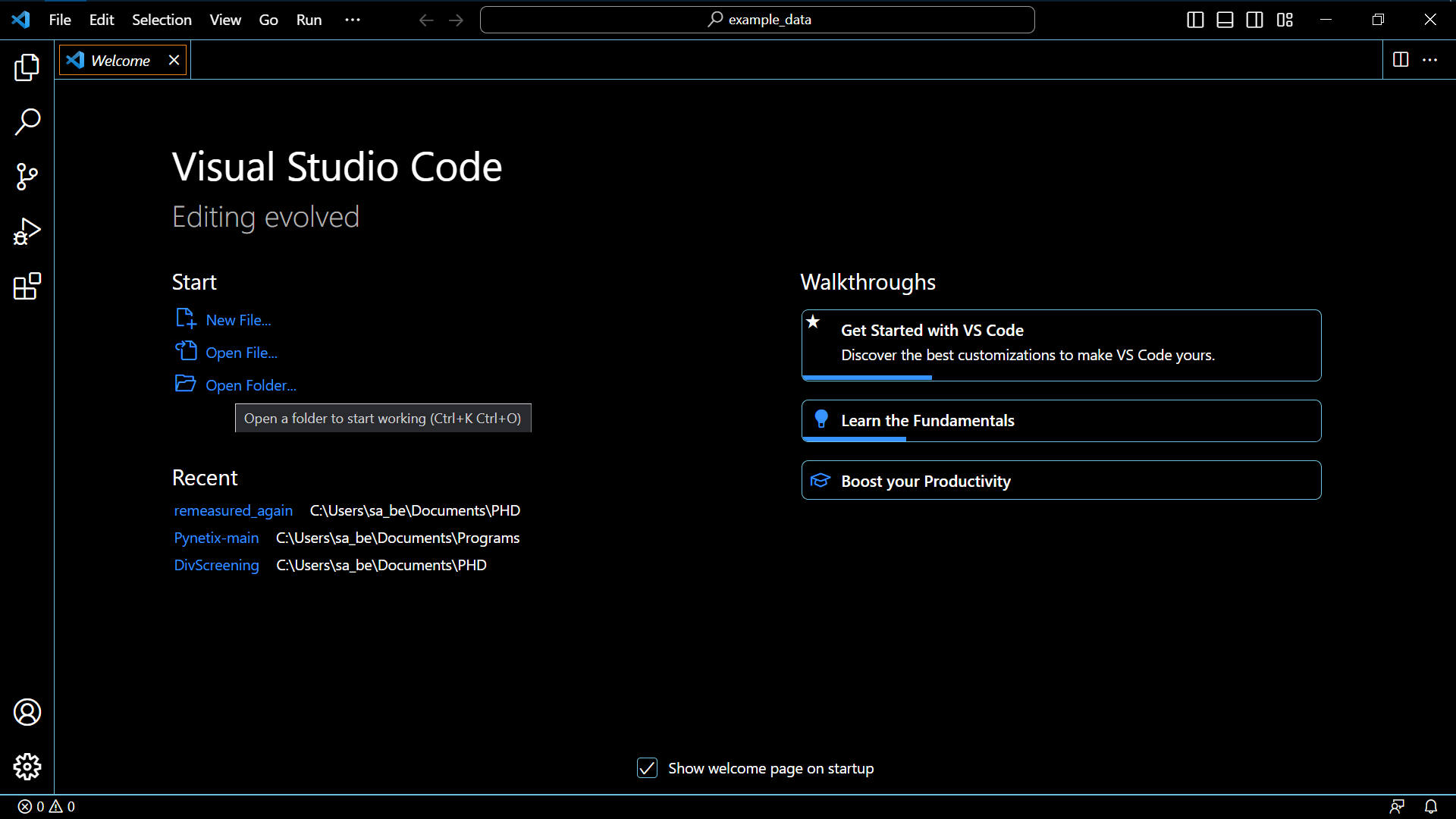
- Open VSC
- Select Open Folder and choose the folder, where your data is stored (here I chose the example folder in this repository)
- Copy the evaluation script (bash-version/evaluation) to this folder
- Open a terminal in VSC
- Be sure it is not a Windows Powershell terminal, but an Ubuntu terminal
- If it is a fresh WSL-Ubuntu install, be sure to run
sudo apt-get update
sudo apt-get upgradethis will make sure, everything will work smoothly. You need to do this just once.
- run the following to install the requirement for the extraction script:
sudo apt-get install poppler-utils
sudo apt-get install rename- Run the script with:
bash ./evaluation- This script works also if multiple folders from different experimental runs are present. For each, an output will be generated. The log-file which you receive as an output can be opened with a simple text-editor.
@article{2023, title={Rapid, Label‐Free Screening of Diverse Biotransformations by Flow‐Injection Mass Spectrometry}, ISSN={1439-7633}, url={http://dx.doi.org/10.1002/cbic.202300170}, DOI={10.1002/cbic.202300170}, journal={ChemBioChem}, publisher={Wiley}, author={Berger, Sarah A.
and Grimm, Christopher and Nyenhuis, Jonathan and Payer, Stefan E. and Oroz-Guinea, Isabel and Schrittwieser, Joerg H. and Kroutil, Wolfgang}, year={2023}, month={Apr} }MIT