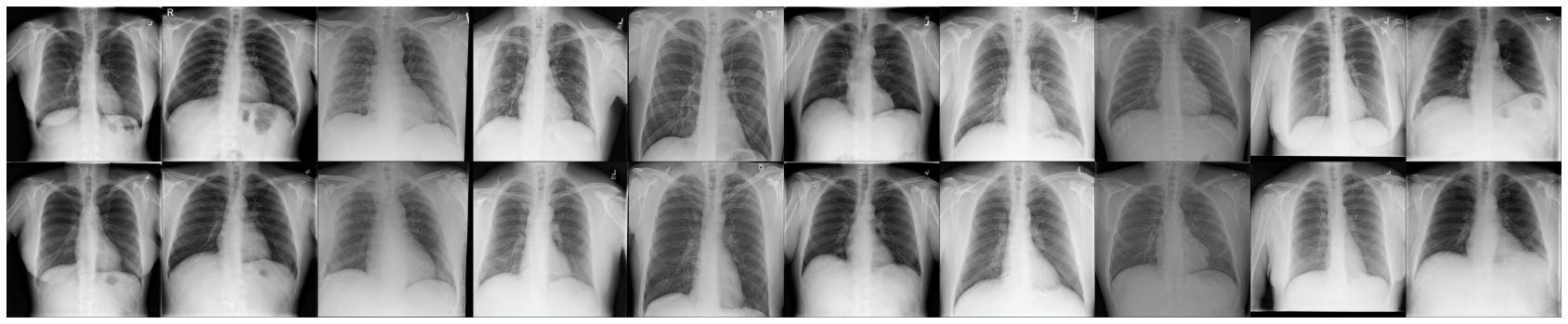
Code for the paper Implicit Embeddings via GAN Inversion for High Resolution Chest Radiographs at the First MICCAI Workshop on Medical Applications with Disentanglements 2022.
Generative models allow for the creation of highly realistic artificial samples, opening up promising applications in medical imaging. In this work, we propose a multi-stage encoder-based approach to invert the generator of a generative adversarial network (GAN) for high resolution chest radiographs. This gives direct access to its implicitly formed latent space, makes generative models more accessible to researchers, and enables to apply generative techniques to actual patient’s images. We investigate various applications for this embedding, including image compression, disentanglement in the encoded dataset, guided image manipulation, and creation of stylized samples. We find that this type of GAN inversion is a promising research direction in the domain of chest radiograph modeling and opens up new ways to combine realistic X-ray sample synthesis with radiological image analysis.
An important building block of our pipeline is the progressive growing generator of Segal et al.. However, it seems that the weights of the generator were removed from their repository and we are not in the position to release it again.
The data used for training is the NIH Chest X-ray 14 dataset, which is available
here. Our default path for the dataset
is assumed to be data/chex-ray14.
Required packages are listed in the requirements.txt file.
The E4E loss uses a pretrained resnet-50 model using mocov2 in the similarity.
It can be downloaded from here.
The default path is assumed to be models/mocov2.pt
The training routines are optimized to work with DDP in a SLURM and will consume all available GPUs in the allocated instance.
The run is configured via the config file configs/inv_boot.yml.
Training is started via:
python scripts/01_train_boot.pyThe run is configured via the config file configs/inv_fine.yml.
Training is started via:
python scripts/02_train_fine.pyHave a look at scripts/03_convert_basic.py to convert the dataset into latent space
using a single encoder pass.
Iterative optimization can be very time and resource consuming. In a lot of cases, the single encoder pass is good enough. The per-sample optimization procedure can be triggered by:
python scripts/04_convert_optim.pyThe repository contains a small showcasing example in notebooks, which illustrates
how to use the model objects.
Both models use convnext_small as backbone.
Please see notebooks/nb_invert_sample.ipynb for an example.
If you use this code for your research, please cite our paper Implicit Embeddings via GAN Inversion for High Resolution Chest Radiographs:
@inproceedings{weber2023implicit,
title={Implicit Embeddings via GAN Inversion for High Resolution Chest Radiographs},
author={Weber, Tobias and Ingrisch, Michael and Bischl, Bernd and R{\"u}gamer, David},
journal={Medical Applications with Disentanglements: First MICCAI Workshop, MAD 2022, Held in Conjunction with MICCAI 2022},
pages={22--32},
year={2023},
organization={Springer}
}