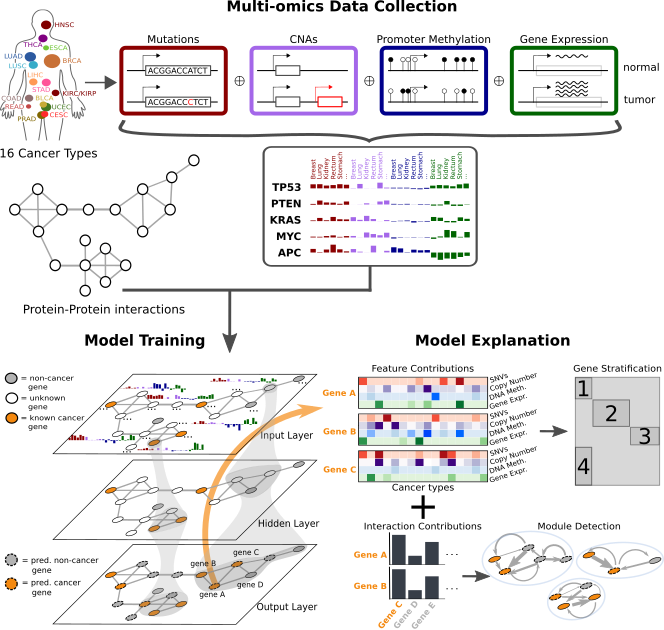
This project predicts cancer genes based on multi-omics feature vectors and protein-protein interactions. Each gene is a data point/node and semi-supervised graph convolutional networks are used for classifying cancer genes. The results surrounding the analysis were published in Nature Machine Intelligence and can be accessed here. The following depicts a broad overview over the EMOGI method.
The analyses from the paper can be entirely reproduced. Several notebooks in the analysis directory allow you to generate all results and the postprocessing script computes ROC and PR curves for EMOGI and competing methods if set up correctly. Please note that you might need the datasets indicated below to properly reproduce all results. To re-train the models, please have a look at the respective section in this README. The existing models also contain the HDF5 containers used for training with fixed training and test splits. You might also want to check out our compute capsule if you are interested in reproducing results, running LRP for your favorite gene or even re-training models without going through the setup.
The code is written in Python 3 and was mainly tested on Python 3.6 and a Linux OS but should run on any OS that supports python and pip. Training is faster on a GPU (which requires the tensorflow-gpu instead of the normal tensorflow package) but works also on a standard computer.
EMOGI has the following dependencies:
- gcn (https://github.com/tkipf/gcn)
- Numpy
- Pandas
- Tensorflow
- h5py
- Networkx
- deepExplain (for an implementation of LRP, https://github.com/marcoancona/DeepExplain)
- mygene (for mapping of gene IDs)
For plotting and correct functioning of all the notebooks, you additionally might need:
- Matplotlib
- Seaborn
- Sklearn
- Scipy
- Goatools (for pathway enrichment)
- UMAP (for dimension reduction)
- UpSetPlot (for visualization of overlaps between datasets/predictions beyond Venn diagrams)
Installing all of the packages should take roughly 10 minutes. To install the gcn package, you have to clone and install the gcn package using:
git clone https://github.com/tkipf/gcn.git
cd gcn
python setup.py install
The other packages can be mostly installed using pip (some are dependenies of others, so no need to install them separately).
Use the following commands to install all required packages (if you don't have root access on your machine, try the --user option of pip):
pip install tensorflow==1.15.3 h5py mygene matplotlib matplotlib-venn seaborn umap-learn goatools UpSetPlot
pip install -e git+https://github.com/marcoancona/DeepExplain.git#egg=deepexplain
To verify a successful setup, you can check out our toy example.
The models that we used in the paper can be downloaded at: http://owww.molgen.mpg.de/~sasse/EMOGI/. Those models can be used to compute LRP contributions and plots like performance comparisons can be done using the postprocessing script in the EMOGI directory.
A trained EMOGI model can be interrogated in a gene-wise fashion to find out why the algorithm considered a gene a cancer gene or not. To compute the feature and interaction partner contributions for a gene of interest, use:
python lrp.py -m <path-to-model-directory> -g <hugo-symbol1> <hugo-symbol2> -b True/False
The genes have to be provided as hugo gene symbols, eg. EGFR or BRCA1. The -b option controls if the resulting plots are heatmaps (more compact, as shown in the paper) or more informative barplots whose error bars indicate standard deviation across cross-validation runs.
Finally, to compute the contributions for all genes (~13,000 to 15,000 depending on the PPI network used), you can specify the -a option. This will take multiple days, however, and uses all available cores.
The output, located in a directory called lrp_sigmoid under the model directory, consists of a pdf file, displaying the multi-omics input feature vector, LRP feature contributions, the most important LRP interaction partner contributions and the multi-omics contributions of the three most important interaction partners for the gene of interest. The multi-omics contributions from the interaction partners are still from the point of view of the gene of interest and indicate that a certain omics feature in a cancer type of an interacting gene was important for the classification of the gene of interest.
A second file contains an edgelist file of interaction partners of the gene of interest, readable in Cytoscape or similar programs.
Run time for a single gene should be around one to two minutes, depending on the number of cores available.
Note: The LRP script is often better not executed on GPU because it doesn't benefit from it and uses a lot of space.
To train EMOGI with your own data, you have to provide a HDF5 container containing the graph, features and labels. There are scripts in pancancer/preprocessing that help with the contruction of the container. In general, a valid container for EMOGI has to contain a graph (called network, a numpy matrix of shape N x N), a feature matrix for the nodes (called features, of shape N x p), the gene names and IDs (called gene_names, as numpy array, dtype object), the training set (called ỳ_train, as boolean indicator array of shape N x 1), the test set of the same shape (called y_test), training and test masks (called train_mask and test_mask, again, indicator arrays of shape N x 1) and the names of the features (called feature_names, an array of length p). Optionally, the raw features can also be added to aid biological interpretation when analyzing the LRP plots (called features_raw, the first row plots the input features and it might be better to plot unnormalized values).
Once you obtained a valid HDF5 container, you can simply train EMOGI with
python train_EMOGI_cv.py -d <path-to-hdf5-container> -hd <n_filters_layer1> <n_filters_layer2>
where the -hd argument specifies the number of graph convolutional layers (the number of arguments) and the number of filters per layer. Training with -hd 100 50 for instance implies that EMOGI is trained with 2 graph convolutional layers with 100 and 50 filters.
In case you want to only train a single model without cross-validation, you can use train_EMOGI.py instead with similar parameters. Use the --help option for an overview of parameters for training.
For the perturbation experiments in the paper, we used the train_all_omics.py script which essentially trains EMOGI models with the same hyper-parameters on a directory of HDF5 containers. See again the help option or the other training scripts for details on the parameters.
To conduct a gridsearch to find suitable combinations of hyper-parameters, use the gridsearch.py script but note, that the combinations are hard-coded into the python file. Other than that, the script should be fairly easy to use. Furthermore, this notebook contains basic analysis to choose a good setting from a successful gridsearch.
To produce basic plots of performance, a prediction for every gene and a comparison with the competing methods, run the postprocessing.py script:
python postprocessing.py -m <path-to-model-directory> -n <name-of-ppi-network> -nm True/False
The name of the PPI network can be any of CPDB, Multinet, PCNet, STRING, IRef or IRefNew and is case-insensitive. The -nm argument specifies if several network measures, such as betweenness centrality, clustering coefficient, degree and others should be included in the comparison to other methods.
Beware that the paths to several datasets are hard-coded in the beginning of the script. If you downloaded these datasets, please correct the paths accordingly.
See the readme file in pancancer for explanations on how you can process your own data and prepare it for EMOGI training.
We used various different data sets from the community to train and validate EMOGI. For deriving positive and negative labels, as well as for functional validation and relationships to other cancer gene sets, a diverse set of data sets were used. Links to the databases and publications can be found below. Ideally, the jupyter notebooks in this repository contain a data section in the beginning where you can exchange the paths with your own copies of the files. Furthermore, some crucial scripts (such as postprocessing and computation of LRP) will work but not produce all of the plots if certain datasets are not available.
We downloaded the file from the preprint. The data is available here: http://ncg.kcl.ac.uk/cancer_genes.php and we downloaded everything together from http://ncg.kcl.ac.uk/download_file.php?file=cancergenes_list.txt
https://cancer.sanger.ac.uk/cosmic/download
https://www.gsea-msigdb.org/gsea/msigdb/cards/KEGG_PATHWAYS_IN_CANCER.html
http://210.107.182.61/digseeOld/
https://ndownloader.figshare.com/files/22629068
https://www.oncokb.org/cancerGenes
http://ongene.bioinfo-minzhao.org/ongene_human.txt
https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp#C4
Table S1 from https://www.cell.com/cms/10.1016/j.cell.2018.02.060/attachment/cf6b14b1-6af1-46c3-a2c2-91008c78e87f/mmc1.xlsx