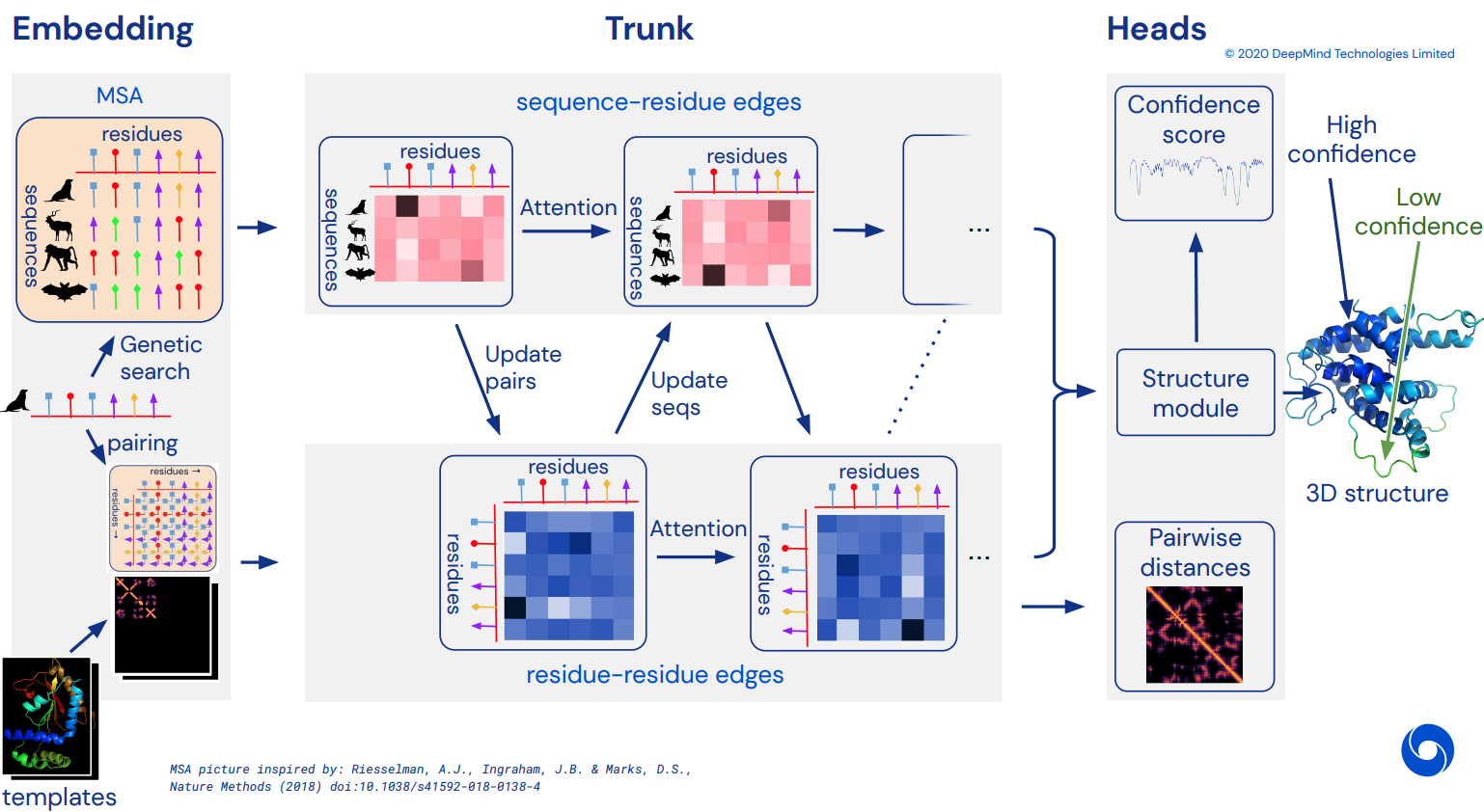
Alphafold2 - Pytorch (wip)
To eventually become an unofficial working Pytorch implementation of Alphafold2. Will be gradually implemented as more details of the architecture is released.
If you are interested in replication efforts, please drop by #alphafold at this Discord channel
Install
$ pip install alphafold2-pytorchUsage
import torch
from alphafold2_pytorch import Alphafold2
model = Alphafold2(
dim = 256,
depth = 2,
heads = 8,
dim_head = 64
).cuda()
seq = torch.randint(0, 21, (1, 128)).cuda()
msa = torch.randint(0, 21, (1, 5, 64)).cuda()
mask = torch.ones_like(seq).bool().cuda()
msa_mask = torch.ones_like(msa).bool().cuda()
distogram = model(
seq,
msa,
mask = mask,
msa_mask = msa_mask
) # (1, 128, 128, 37)Data
This library will use the awesome work by Jonathan King at this repository.
To install
$ git clone https://github.com/jonathanking/sidechainnet.git
$ cd sidechainnet && pip install -e .Speculation
https://fabianfuchsml.github.io/alphafold2/
Developments from competing labs
https://www.biorxiv.org/content/10.1101/2020.12.10.419994v1.full.pdf
Final step - Fast Relax
Citations
@misc{unpublished2021alphafold2,
title={Alphafold2},
author={John Jumper},
year={2020},
archivePrefix={arXiv},
primaryClass={q-bio.BM}
}@misc{king2020sidechainnet,
title={SidechainNet: An All-Atom Protein Structure Dataset for Machine Learning},
author={Jonathan E. King and David Ryan Koes},
year={2020},
eprint={2010.08162},
archivePrefix={arXiv},
primaryClass={q-bio.BM}
}@misc{alquraishi2019proteinnet,
title={ProteinNet: a standardized data set for machine learning of protein structure},
author={Mohammed AlQuraishi},
year={2019},
eprint={1902.00249},
archivePrefix={arXiv},
primaryClass={q-bio.BM}
}