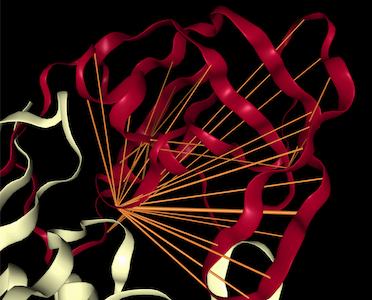
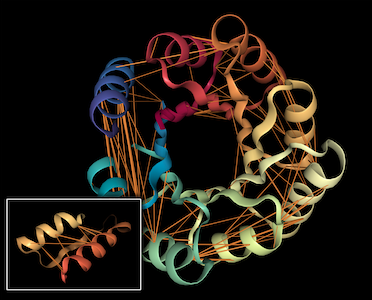
This repository is the official implementation of BERTology Meets Biology: Interpreting Attention in Protein Language Models.
This section provides instructions for generating visualizations of attention projected onto 3D protein structure.
General requirements:
- Python >= 3.7
pip install biopython==1.77
pip install tape-proteins==0.4
pip install jupyterlab==3.0.14
pip install nglview
jupyter-nbextension enable nglview --py --sys-prefix
If you run into problems installing nglview, please refer to their installation instructions for additional installation details and options.
cd <project_root>/notebooks
jupyter notebook provis.ipynb
If you get an error running the notebook, you may need to execute the notebook as follows:
jupyter notebook --NotebookApp.iopub_data_rate_limit=10000000
See nglview installation instructions for more details.
You may edit the notebook to choose other proteins, attention heads, etc. The visualization tool is based on the excellent nglview library.
This section describes how to reproduce the experiments in the paper.
cd <project_root>
python setup.py develop
To download additional required datasets from TAPE:
cd <project_root>/data
wget http://s3.amazonaws.com/proteindata/data_pytorch/secondary_structure.tar.gz
tar -xvf secondary_structure.tar.gz && rm secondary_structure.tar.gz
wget http://s3.amazonaws.com/proteindata/data_pytorch/proteinnet.tar.gz
tar -xvf proteinnet.tar.gz && rm proteinnet.tar.gz
The following steps will reproduce the attention analysis experiments and generate the reports currently found in <project_root>/reports/attention_analysis. This includes all experiments besides the probing experiments (see Probing Analysis).
Before performing steps, navigate to appropriate directory:
cd <project_root>/protein_attention/attention_analysis
The following executes the attention analysis (may run for several hours):
sh scripts/compute_all_features_tape_bert.sh
The above script create a set of extract files in <project_root>/data/cache corresponding to various properties
being analyzed. You may edit the script files to remove properties that you are not interested in. If you wish to run the
analysis without a GPU, you must specify the --no_cuda flag.
The following generate reports based on the files created in previous step:
sh scripts/report_all_features_tape_bert.sh
If you removed steps from the analysis script above, you will need to update the reporting script accordingly.
In order to generate reports for the ProtTrans models, follow the instructions as for the TapeBert
model above, but substitute the following commands:
ProtBert:
sh scripts/compute_all_features_prot_bert.sh
sh scripts/report_all_features_prot_bert.sh
ProtBertBFD:
sh scripts/compute_all_features_prot_bert_bfd.sh
sh scripts/report_all_features_prot_bert_bfd.sh
ProtAlbert:
sh scripts/compute_all_features_prot_albert.sh
sh scripts/report_all_features_prot_albert.sh
ProtXLNet:
sh scripts/compute_all_features_prot_xlnet.sh
sh scripts/report_all_features_prot_xlnet.sh
The following steps will recreate the figures from the probing analysis, currently found in <project_root>/reports/probing
Navigate to directory:
cd <project_root>/protein_attention/probing
Train diagnostic classifiers. Each script will write out an extract file with evaluation results. Note: each of these scripts may run for several hours.
sh scripts/probe_ss4_0_all
sh scripts/probe_ss4_1_all
sh scripts/probe_ss4_2_all
sh scripts/probe_sites.sh
sh scripts/probe_contacts.sh
python report.py
This project is licensed under BSD3 License - see the LICENSE file for details
This project incorporates code from the following repo:
When referencing this repository, please cite this paper.
@misc{vig2020bertology,
title={BERTology Meets Biology: Interpreting Attention in Protein Language Models},
author={Jesse Vig and Ali Madani and Lav R. Varshney and Caiming Xiong and Richard Socher and Nazneen Fatema Rajani},
year={2020},
eprint={2006.15222},
archivePrefix={arXiv},
primaryClass={cs.CL},
url={https://arxiv.org/abs/2006.15222}
}