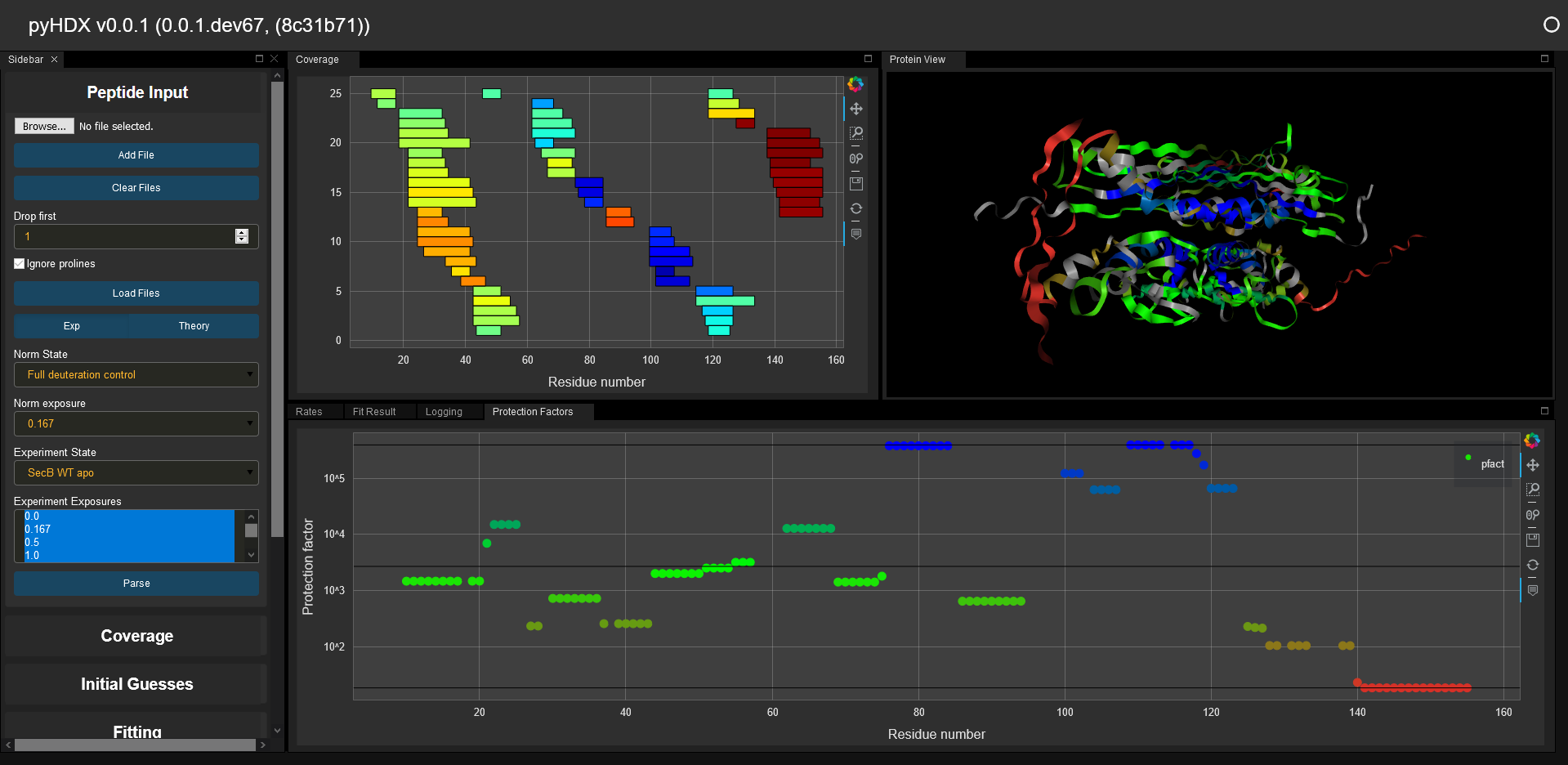
PyHDX is python project which can be used to derive Gibbs free energy from HDX-MS data. Currently the project is functional but in beta, future versions will likely subject to changes in API and analysis.
Installation with conda:
(Currently PyHDX 0.3.2 is unavailable on conda-forge due to issues with python 3.9 please use 0.3.0 or install from source/pip)
$ conda install -c conda-forge pyhdx
Installation with pip:
$ pip install pyhdx
Development install instructures in docs/installation.rst
$ pyhdx serve
Please refer to the docs for more details on how to run PyHDX.
The PyHDX web application is currently hosted at: http://pyhdx.jhsmit.org/main
A test file can be downloaded from here and here. (right click, save as).
Two other web applications are available. To upload fitting results from the main application and visualize: http://pyhdx.jhsmit.org/single
To upload multiple fitting result datasets and compare and visualize: http://pyhdx.jhsmit.org/diff
The 0.4.0 beta version of PyHDX (featuring batch fitting / multiple states) is hosted at: http://pyhdx-beta.jhsmit.org/main