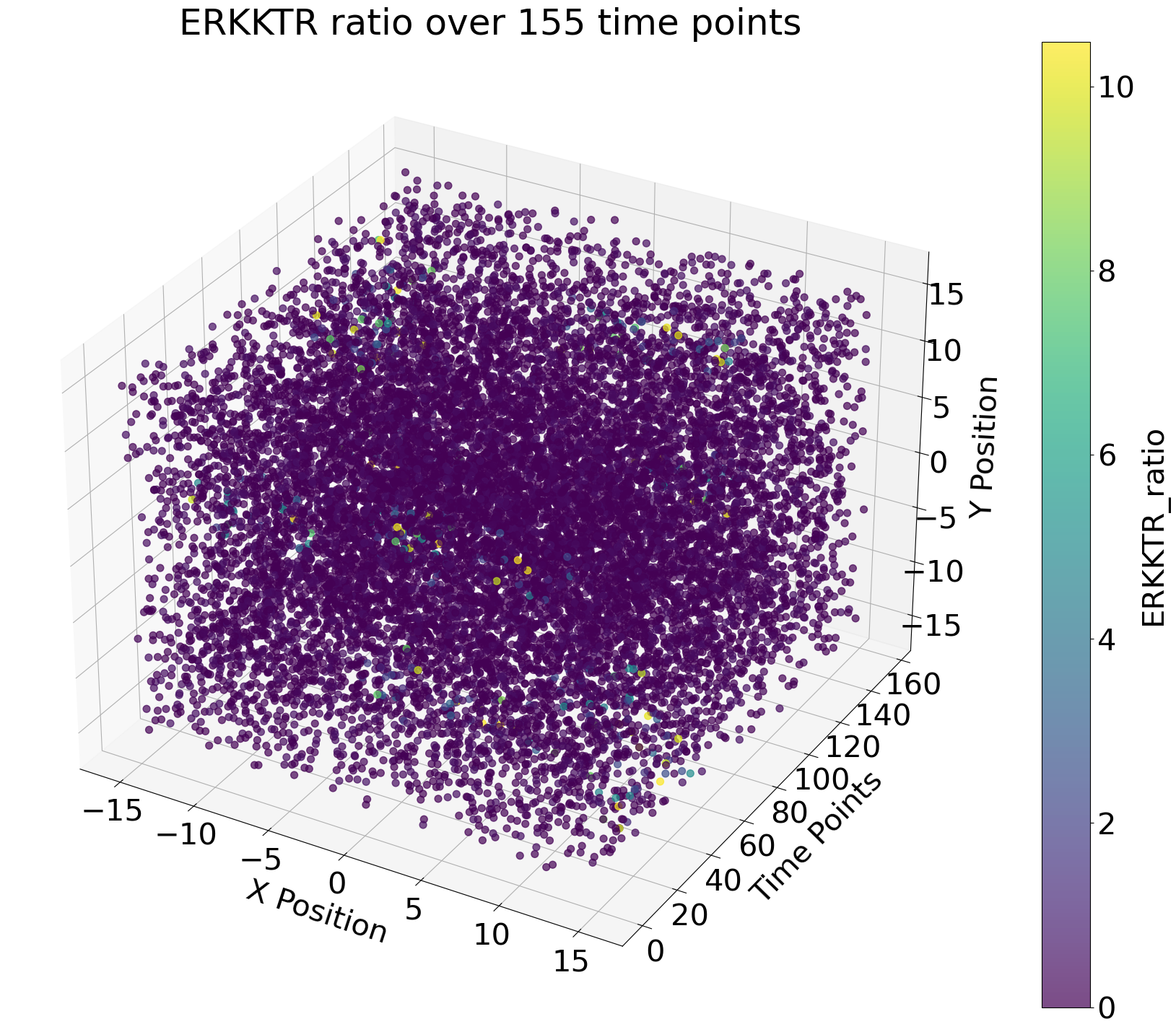
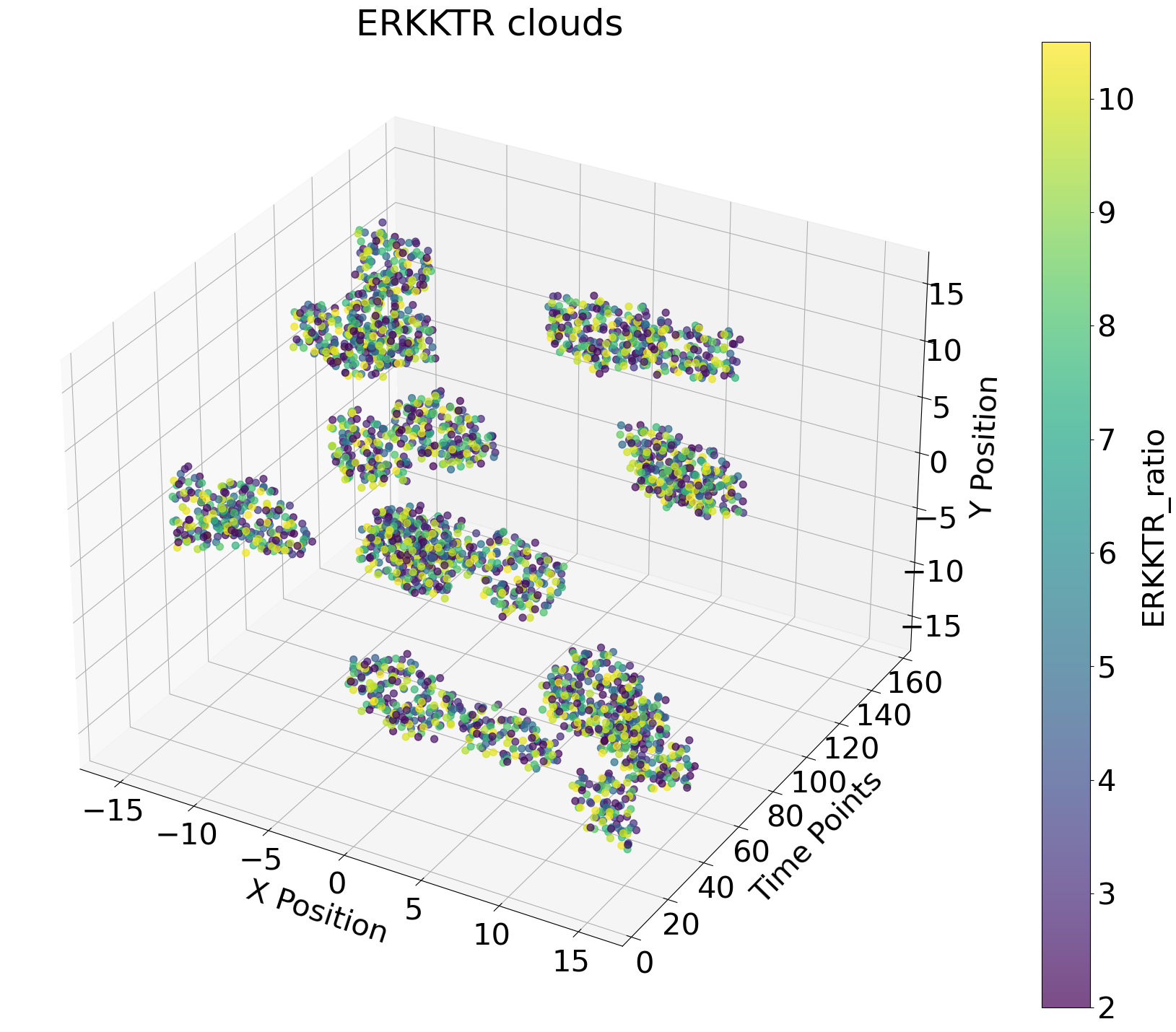
This project is part of my masters thesis in computational biology in collaboration with the Cellular Signalling lab of Prof. Olivier Pertz at the University of Bern. We aim to create a neural network that can reliably identify intercellular signals in cancer epithelium over time. Instead of using traditional image analysis, we are tyring to reduce the data size by representing the cells in the tissue over time as points in a point cloud. In the cloud, the x- and y-axis are the coordinates of the cell in the original microscopy image and the z-axis represents time.
After having started with a box detection system, we have now switched to instance segmentation for each cell only. The boxes require way more computational power than instance segmentation alone. Although, the boxes would make the network prediction more reliable, as each instance of a signalling wave is also given by a box and not just a mask, it is also not necessary when a model is trained well enough.
The box detection can be found in the source directory, and the instance segmentation in the source_inst one. Both use the pytorch geometric framework for point cloud representation and the correcponding neural network components.