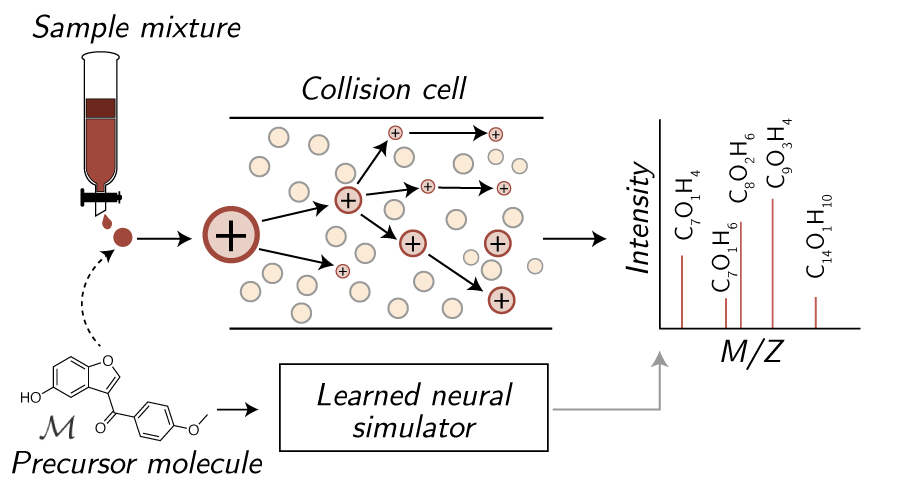
This repository contains implementations for the following spectrum simulator models predicting molecular tandem mass spectra from molecules:
- ️️️️ ❄️ ICEBER️️G ❄️: Inferring CID by Estimating Breakage Events and Reconstructing their Graphs (now available to run through GNPS2 as a dropdown option within the 3DMolMS workflow)
- 🧣 SCARF 🧣: Subformula Classification for Autoregressively Reconstructing Fragmentations,
ICEBERG predicts spectra at the level of molecular fragments, whereas SCARF predicts spectra at the level of chemical formula. In order to fairly compare various spectra models, we implement a number of baselines and alternative models using equivalent settings across models (i.e., same covariates, hyeprparmaeter sweeps for each, etc.):
- NEIMS using both FFN and GNN encoders from Rapid prediction of electron–ionization mass spectrometry using neural networks
- MassFormer from MassFormer: Tandem Mass Spectrum Prediction for Small Molecules using Graph Transformers
- 3DMolMS from 3DMolMS: Prediction of Tandem Mass Spectra from Three Dimensional Molecular Conformations
- GRAFF-MS from Efficiently predicting high resolution mass spectra with graph neural networks
- CFM-ID from CFM-ID 4.0: More Accurate ESI-MS/MS Spectral Prediction and Compound Identification (not retrained; instructions for running are provided)
Contributors: Sam Goldman, John Bradshaw, Janet Li, Jiayi Xin, Connor W. Coley
Install and set up the conda environment using mamba:
mamba env create -f environment.yml
mamba activate ms-gen
pip install -r requirements.txt
python3 setup.py develop
Note: if you are using GPU, please uncomment the CUDA-based packages for DGL and comment the CPU package in envorinment.yaml.
To make predictions, we have released and made public a version of SCARF and ICEBERG both trained upon the CANOPUS dataset (renamed NPLIB1 for clarity in corresponding manuscripts). This can be downloaded and used to predict a set of 100 sample molecules contained in the NIST library, as included at data/spec_datasets/sample_labels.tsv (test set):
# Scarf quickstart
. quickstart/scarf/download_model.sh
. quickstart/scarf/run_model.sh
# Iceberg quickstart
. quickstart/iceberg/download_model.sh
. quickstart/iceberg/run_model.sh
Model outputs will be contained in quickstart/{model}/out/. We note that this model may be less performant than the model trained on the commercial NIST20 Library. Download links to models trained on NIST20 models are available upon request to any users with a NIST license. ICEBERG assigns intensities to various fragment masses. Refer to notebooks/scarf_demo.ipynb and notebooks/iceberg_demo.ipynb for a walkthrough on how to process and interpret the output dictionary.
Please note models have been updated since the original release to correct for a small error in the adduct masses for [M-H2O+H]+ and should be re-downloaded.
A data subset from the GNPS database including processed dag annotations (magma_outputs/), subformulae (subformulae/), retrieval databases (retrieval/), splits (splits/), and spectra files (spec_files/) can be downloaded into data/spec_datasets/canopus_train_public. Note CANOPUS is an earlier name of the GNPS dataset, as explained in our ICEBERG paper.
. data_scripts/download_gnps.sh
For those interested in understanding the full dataset processing pipeline, we refer you to data_scripts/. This contains functinoalities for generating assignment DAGs (i.e., iceberg training), assigning subformulae to spectra (i.e., scarf training + evaluation), and pubchem dataset creation (i.e., for retrieval experiments). We explain some of these details below.
nist20 is a commercial dataset. Many of the scripts and pipelines include commands to run / train models on NIST20 as well; we suggest commenting these out where applicable.
As part of revisions for our work introducing ICEBERG, we also evaluated our pretrained models on the CASMI22 positive mode spectra test set. We extracted CASMI22 previously and show how to post-process and download this data in the notebook notebooks/iceberg_casmi22.ipynb.
Data should then be assigned to subformulae files using
data_scripts/forms/assign_subformulae.py, which will preprocess the data. We
produce two fragment versions of the molecule, magma_subform_50 and
no_subform. The former strictly labels subformula based upon smiles structure
and the latter is permissive and allows all entries to pass. The following
script does this for both canopus_train_public and nist20 if it has been
acquired and parsed (commercial dataset).
. data_scripts/all_assign_subform.sh
In addition to building processed subformulae, to train ICEBERG, we must annotate substructures and create a labeled dataset over the breakage process, which we do with the MAGMa algorithm.
This can be done with the following script, specifying an appropriate dataset:
. data_scripts/dag/run_magma.sh
To conduct retrieval experiments, libraries of smiles must be created. A PubChem library is converted and each chemical formula is mapped to (smiles, inchikey) pairs. Subsets are selected for evaluation. Making formula subsets takes longer (on the order of several hours, even parallelized) as it requires converting each molecule in pubchem to a mol / InChi.
source data_scripts/pubchem/01_download_smiles.sh
python data_scripts/pubchem/02_make_formula_subsets.py
python data_scripts/pubchem/03_dataset_subset.py --dataset-labels data/spec_datasets/nist20/labels.tsv # for nist20 dataset
python data_scripts/pubchem/04_make_retrieval_lists.py
Processed tables are already included inside canopus_train_public.
SCARF models trained in two parts: a prefix tree generator and an intensity predictor. The pipeline for training and evaluating this model can be accessed in run_scripts/scarf_model/. The internal pipeline used to conduct experiments can be followed below:
- Train scarf model:
run_scripts/scarf_model/01_run_scarf_gen_train.sh - Sweep number of prefixes to generate:
run_scripts/scarf_model/02_sweep_scarf_gen_thresh.py - Use model 1 to predict model 2 training set:
run_scripts/scarf_model/03_scarf_gen_predict.sh - Train intensity model:
run_scripts/scarf_model/04_train_scarf_inten.sh - Make and evaluate intensity predictions:
run_scripts/scarf_model/05_predict_form_inten.py - Run retrieval:
run_scripts/scarf_model/06_run_retrieval.py - Time scarf:
run_scripts/scarf_model/07_time_scarf.py - Export scarf forms
run_scripts/scarf_model/08_export_forms.py
Instead of running in batched pipeline model, individual gen training, inten training, and predict calls can be made using the following scripts respectively:
python src/ms_pred/scarf_pred/train_gen.pypython src/ms_pred/scarf_pred/train_inten.pypython src/ms_pred/scarf_pred/predict_smis.py
An additional notebook showcasing how to individually load models and make predictions can be found at notebooks/scarf_demo.ipynb.
We provide scripts showing how we conducted hyperparameter optimization as well:
- Hyperopt scarf model:
run_scripts/scarf_model/hyperopt_01_scarf.sh - Hyperopt scarf inten model:
run_scripts/scarf_model/02_sweep_scarf_gen_thresh.py
ICEBRG models, like SCARF, are trained in two parts: a learned fragment generator and an intensity predictor. The pipeline for training and evaluating this model can be accessed in run_scripts/dag_model/. The internal pipeline used to conduct experiments can be followed below:
- Train dag model:
run_scripts/dag_model/01_run_dag_gen_train.sh - Sweep over the number of fragments to generate:
run_scripts/dag_model/02_sweep_gen_thresh.py - Use model 1 to predict model 2 training set:
run_scripts/dag_model/03_run_dag_gen_predict.sh - Train intensity model:
run_scripts/dag_model/04_train_dag_inten.sh - Make and evaluate intensity predictions:
run_scripts/dag_model/05_predict_dag_inten.py - Run retrieval:
run_scripts/dag_model/06_run_retrieval.py - Time iceberg:
run_scripts/dag_model/07_time_dag.py - Export dag predictions
run_scripts/dag_model/08_export_preds.py
Instead of running in batched pipeline model, individual gen training, inten training, and predict calls can be made using the following scripts respectively:
python src/ms_pred/dag_pred/train_gen.pypython src/ms_pred/dag_pred/train_inten.pypython src/ms_pred/dag_pred/predict_smis.py
An additional notebook showcasing how to individually load models and make predictions can be found at notebooks/iceberg_demo.ipynb.
The models were hyperoptimized using the following scripts:
run_scripts/dag_model/hyperopt_01_dag.shrun_scripts/dag_model/hyperopt_02_inten.sh
Experiment pipeline utilized:
- Train models:
run_scripts/ffn_model/01_run_ffn_train.sh - Predict and eval:
run_scripts/ffn_model/02_predict_ffn.py - Retreival experiments:
run_scripts/ffn_model/03_run_retrieval.py - Time ffn:
run_scripts/ffn_model/04_time_ffn.py
Hyperopt FFN: run_scripts/ffn_model/hyperopt_01_ffn.sh
Baseline used to show the effect of successively generating formula, rather than decoding with SCARF.
Experiment pipeline utilized:
- Train models:
run_scripts/autoregr_baseline/01_run_autoregr_train.sh - Sweep model:
run_scripts/autoregr_baseline/02_sweep_autoregr_thresh.py
Hyperparameter optimization: run_scripts/autoregr_baseline/hyperopt_01_autoregr.sh
Experiment pipeline:
- Train models:
run_scripts/gnn_model/01_run_gnn_train.sh - Predict and eval:
run_scripts/gnn_model/02_predict_gnn.py - Retreival experiments:
run_scripts/gnn_model/03_run_retrieval.py - Time gnn:
run_scripts/gnn_model/04_time_gnn.py
Hyperopt GNN: run_scripts/gnn_model/hyperopt_01_gnn.sh
Experiment pipeline:
- Train models:
run_scripts/massformer_model/01_run_massformer_train.sh - Predict and eval:
run_scripts/massformer_model/02_predict_massformer.py - Retreival experiments:
run_scripts/massformer_model/03_run_retrieval.py - Time massformer:
run_scripts/massformer_model/04_time_massformer.py
Hyperopt Massformer: run_scripts/massformer_model/hyperopt_01_massformer.sh
We include a baseline implementation of 3DMolMS in which we utilize the same architecture as these authors. We note we do not include collision energy or machines as covariates for consistency with our other implemented models and data processing pipelines, which may affect performance.
Experiment pipeline:
- Train models:
run_scripts/molnetms/01_run_ffn_train.sh - Predict and eval:
run_scripts/molnetms/02_predict_ffn.py - Retreival experiments:
run_scripts/molnetms/03_run_retrieval.py - Time 3d mol ms:
run_scripts/molnetms/04_time_molnetms.py
Hyperopt 3DMolMS: run_scripts/molnetms/hyperopt_01_molnetms.sh
We include a baseline variation of GRAFF-MS in which we utilize a fixed formula vocabulary. We note we do not include collision energy or machines as covariates for consistency with our other implemented models and data processing pipelines, which may affect performance. In addition, because our data does not contain isotopic or varied adduct formula labels, we replace the marginal peak loss with a cosine similarity loss. Pleases see the original paper to better understand the release details.
Experiment pipeline:
- Train models:
run_scripts/graff_ms/01_run_ffn_train.sh - Predict and eval:
run_scripts/graff_ms/02_predict_ffn.py - Retreival experiments:
run_scripts/graff_ms/03_run_retrieval.py - Time graff MS:
run_scripts/graff_ms/04_time_graff_ms.py
Hyperopt graff ms: run_scripts/graff_ms/hyperopt_01_graff_ms.sh
CFM-ID is a well-established fragmentation-based mass spectra prediction model. We include brief instructions for utilizing this tool below
Build docker:
docker pull wishartlab/cfmid:latest
Make prediction:
. run_scripts/cfm_id/run_cfm_id.py
. run_scripts/cfm_id/process_cfm.py
. run_scripts/cfm_id/process_cfm_pred.py
As an addiitonal baseline to compare to the generative portion of our scarf (thread), we include frequency baselines for generating form subsets:
. run_scripts/freq_baseline/predict_freq.py
. run_scripts/freq_baseline/predict_rand.py
Analysis scripts can be found in analysis for evaluating both formula
predictios analysis/form_pred_eval.py and spectra predictions
analysis/spec_pred_eval.py.
Additional analyses used for figure generation were conducted in notebooks/.
A common use case for forward spectrum prediction models is to use the trained model as a surrogate model for augmenting an inverse model (e.g., MIST). An example workflow for doing this is shown in run_scripts/iceberg_augmentation. The target dataset including a labels file, spectrum files, split, and target augmetnation file for prediction should first be coppied into the data/spec_datasets. Once this is complete, the runscripts folder can be copied, modified to use the datset of interest, and run. The ideal output will be a single MGF and labels files including the ouptut predictions.
We ask any user of this repository to cite the following works based upon the portion of the repository used:
@article{https://doi.org/10.48550/arxiv.2303.06470,
doi = {10.48550/ARXIV.2303.06470},
url = {https://arxiv.org/abs/2303.06470},
author = {Goldman, Samuel and Bradshaw, John and Xin, Jiayi and Coley, Connor W.},
keywords = {Quantitative Methods (q-bio.QM), Machine Learning (cs.LG), FOS: Biological sciences, FOS: Biological sciences, FOS: Computer and information sciences, FOS: Computer and information sciences},
title = {Prefix-tree Decoding for Predicting Mass Spectra from Molecules},
publisher = {arXiv},
year = {2023},
copyright = {Creative Commons Attribution Share Alike 4.0 International}
}
@misc{goldman2023generating,
title={Generating Molecular Fragmentation Graphs with Autoregressive Neural Networks},
author={Samuel Goldman and Janet Li and Connor W. Coley},
year={2023},
eprint={2304.13136},
archivePrefix={arXiv},
primaryClass={q-bio.QM}
}
In addition, we utilize both the NEIMS approach for our binned FFN and GNN encoders, 3DMolMS, GRAFF-MS, Massformer, MAGMa for constructing formula labels, and CFM-ID as a baseline. We encourage considering the following additional citations:
- Wei, Jennifer N., et al. "Rapid prediction of electron–ionization mass spectrometry using neural networks." ACS central science 5.4 (2019): 700-708.
- Ridder, Lars, Justin JJ van der Hooft, and Stefan Verhoeven. "Automatic compound annotation from mass spectrometry data using MAGMa." Mass Spectrometry 3.Special_Issue_2 (2014): S0033-S0033.
- Wang, Fei, et al. "CFM-ID 4.0: more accurate ESI-MS/MS spectral prediction and compound identification." Analytical chemistry 93.34 (2021): 11692-11700.
- Hong, Yuhui, et al. "3DMolMS: Prediction of Tandem Mass Spectra from Three Dimensional Molecular Conformations." bioRxiv (2023): 2023-03.
- Murphy, Michael, et al. "Efficiently predicting high resolution mass spectra with graph neural networks." arXiv preprint arXiv:2301.11419 (2023).
- Young, Adamo, Bo Wang, and Hannes Röst. "MassFormer: Tandem mass spectrum prediction with graph transformers." arXiv preprint arXiv:2111.04824 (2021).