This repository contains code to train a single Convolutional Neural Network to segment MRI scans of different contrasts
and resolutions. The network is trained with synthetic scans obtained by sampling a generative model building on the
lab2im package, which we really encourage you to have a look at!
This project is explained in details in the following video:

In short, synthetic scans are generated by sampling a Gaussian Mixture Model (GMM) conditioned
on training label maps. The variability of the generated images is further extended by performing data augmentation
steps such as spatial deformation, intensity augmentation, and random blurring. The following figure illustrates some
of the many generation possibilities offered by SynthSeg:
realistic/unrealistic, isotropic/anisotropic, uni-modal/multi-modal scans.
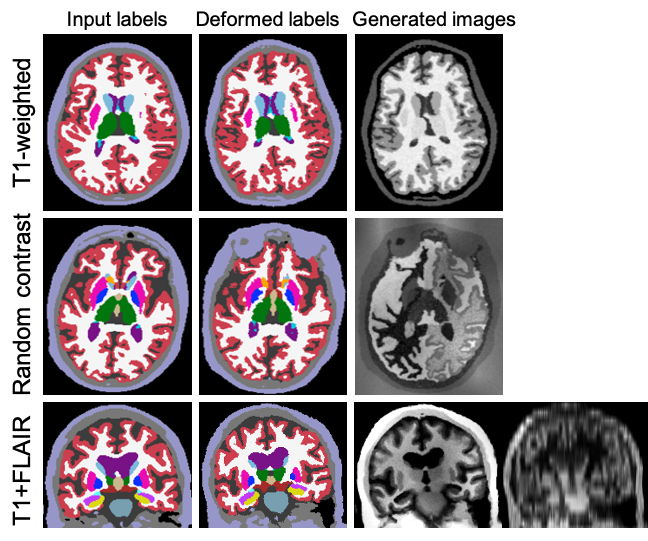
The generated images are then usd to train a CNN for image segmentation. Because the parameters of the GMM (means and
variances) are sampled at each minibatch from prior distributions, the network is exposed to images yielding differences
in contrast (depending on the GMM priors), and learns robust features. An overvoew of the training process is
represented in the following figure:
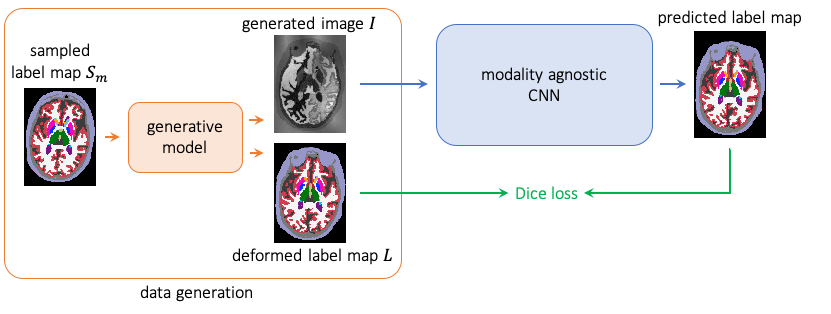
Additionally to the training function, we provide the generative model labels_to_image_model, along with a wrapper
brain_generator, which allows to easily synthesise new images. This repository also contains functions for evaluation,
validation (which must be done offline), and prediction.
If you wish to train your own network or try the generative model, you can familiarise yourself with the different parameters by trying the provided scripts, classified in three folders:
-
tutorials: We advise you to start here with the three simple examples of this folder. simple_example.py shows how to generate images in three lines with the
BrainGeneratorwrapper. Then random_contrst_generation.py and t1w_generation.py introduce some basic parameters to easily generate images of random or constrained intenisty distributions. -
SynthSeg_scripts: this folder contains two scripts showing how we trained SynthSeg, a contrast-agnostic network for segmenation of brain MRI scans. These scripts introduce some new parameters of the generative model.
-
PV-SynthSeg_scripts: this folder contains several scripts, among which you can find how we trained PV-SynthSeg, a PV-aware network for segmentation of brain MRI scans. In particular the PV-SynthSeg_generation explains some advanced parameters if you wish to generate multi-modal and/or anisotropic images.
-
SynthSeg: this is the main folder containing the generative model and training function:
-
labels_to_image_model.py: contains the generative model
labels_to_image_model. -
brain_generator.py: contains the class
BrainGenerator, which is a wrapper aroundlabels_to_image_model. New images can simply be generated by instantiating an object of this class, and call the methodgenerate_image(). -
model_inputs.py: contains the function
build_model_inputsthat prepares the inputs of the generative model. -
training.py: contains the function
trainingto train the segmentation network. This function provides an example of how to integrate the labels_to_image_model in a broader GPU model. All training parameters are explained there. -
metrics_model.py: contains a keras model that implements the computation of a Soft Dice loss function.
-
predict.py: function to predict segmentations of new MRI scans. Can also be used for testing.
-
evaluate.py: contains functions to evaluate the accuracy of segmentations with several metrics:
dice_evaluation,surface_distances, andcompute_non_parametric_paired_test. -
validate.py: contains
validate_trainingto validate the models saved during training, as validation on real images has to be done offline. -
supervised_training.py: contains all the functions necessary to train a model on real MRI scans.
-
estimate_priors.py: contains functions to estimate the prior distributions of the GMM parameters.
-
-
script: additionally to the tutorials, we also provide functions such as to launch your own trainings.
-
ext: contains external packages, especially the lab2im package, and a modified version of neuron.
This code relies on several external packages (already included in \ext):
-
lab2im: contains functions for data augmentation, and a simple version of the generative model, on which we build to build
label_to_image_model -
neuron: contains functions for deforming, and resizing tensors, as well as functions to build the segmentation network [1,2].
-
pytool-lib: library required by the neuron package.
All the other requirements are listed in requirements.txt. We list here the important dependencies:
- tensorflow-gpu 2.0
- tensorflow_probability 0.8
- keras > 2.0
- cuda 10.0 (required by tensorflow)
- cudnn 7.0
- nibabel
- numpy, scipy, sklearn, tqdm, pillow, matplotlib, ipython, ...
If you use this code, please cite the following papers:
A Learning Strategy for Contrast-agnostic MRI Segmentation
Benjamin Billot, Douglas N. Greve, Koen Van Leemput, Bruce Fischl, Juan Eugenio Iglesias*, Adrian V. Dalca*
*contributed equally
accepted for MIDL 2020
[ arxiv | bibtex ]
Partial Volume Segmentation of Brain MRI Scans of any Resolution and Contrast
Benjamin Billot, Eleanor D. Robinson, Adrian V. Dalca, Juan Eugenio Iglesias
under revision
[ arxiv | bibtex ]
If you have any question regarding the usage of this code, or any suggestions to improve it you can contact me at: benjamin.billot.18@ucl.ac.uk
[1] Anatomical Priors in Convolutional Networks for Unsupervised Biomedical Segmentation Adrian V. Dalca, John Guttag, Mert R. Sabuncu, 2018
[2] Unsupervised Data Imputation via Variational Inference of Deep Subspaces Adrian V. Dalca, John Guttag, Mert R. Sabuncu, 2019