galen-view is a visualizer for exploring the CORD-19 dataset on a stand-alone desktop computer. The code is intended to be small and straight-forward, enabling researchers to expand and modify the code.
This software was written as part of an effort at Sandia National Laboratories described here
This code is named after Galen, a second century Greek physician who documented the Antonine Plague in the Roman Empire.
galen-view was written and tested in python 3 and uses syntax specific to python 3. To get started, create a python 3 environment.
For example, if you are using anaconda, you can execute the following:
conda create -n galenenv python=3
conda activate galenenvIf you are using Anaconda on Windows 10, also type:
conda install holoviewsInstalling the wheel file will install galen-view along with it's various dependencies. galen-view is based on holoviews, so there are a large number of dependencies.
This can be done using:
pip install galen-viewThe module sandia.galen.dataprep contains several methods for downloading, analyzing, and indexing the cord-19 dataset. For convenience, the module can be executed as an "all-in-one" call do perform all these funcions and prepare the data for galen-view.
If you are behind a proxy, you will need to set the environment variables in order to download the data. This can be accomplished by setting the http_proxy and https_proxy environment variables. If the "pip install" step above worked, it is highly likely that you already have them set properly. Note that this is only necessary for the download and not for galen-view to work after that.
This will download files and create more. It is recommended to do this in a fresh directory.
To create a new directory:
mkdir galen_data
cd galen_data
Then run one of the following two commands:
For a faster startup time that only analyzes the first 2000 documents (You may have to use "python" rather than "python3"):
python3 -m sandia.galen.dataprep testTo analyze all the downloaded documents, execute the following.
(You may have to use "python" rather than "python3").
This can take hourse, but you only have to do it once:
python3 -m sandia.galen.dataprepYou will see progress during each step. You should see something similar to the following:
$ python3 -m sandia.galen.dataprep
Initializing Data in Current Directory
Checking for Data in this Directory
Downloading the metadata file
Downloading the CORD-19 Dataset
Processing https://path/to/biorxiv_medrxiv.tar.gz ... Done.
Processing https://path/to/comm_use_subset.tar.gz ... Done.
Processing https://path/to/noncomm_use_subset.tar.gz ... Done.
Processing https://path/to/pmc_custom_license.tar.gz ... Done.
Processing https:///custom_license.tar.gz ... Done.
Processing https:///cord19_specter_embeddings_2020-04-03.tar.gz ... Done.
Processing https:///cord19_specter_embeddings_2020-04-10.tar.gz ... Done.
========
Making the Coordinates to Visualize the Data
========
Building a searchable index from the documents
..........
..........
..........
[There will be more lines like this]
..........
..........
..........
Done. Committing Index
Done Indexing
========
Done. Run "jupyter-notebook GalenView.ipynb" and run all cells to explore the data.
sandia.galen.dataprep's final step is to copy a jupyter notebook into the directory that can be used to view and explore the data. You can open this notebook in a variety of ways, but the following will open it directly:
jupyter-notebook GalenView.ipynbAfter executing this, a web browser should open with the jupyter notebook. You still have to run all the cells in the notebook to activate and use the visualization.
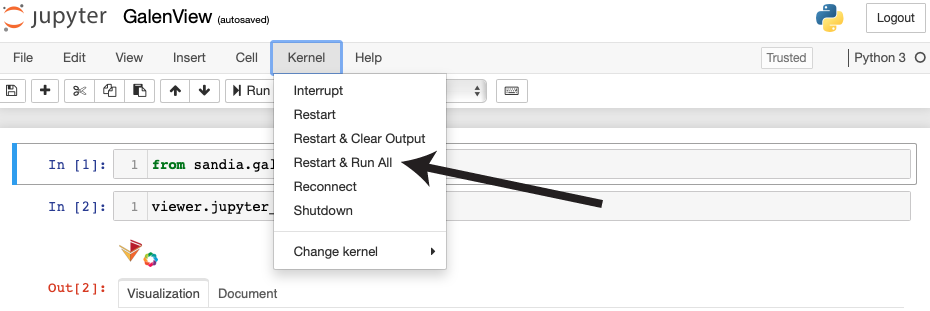
| After the notebook loads, click "Restart & Runn All" from the menu at the top of the page. |
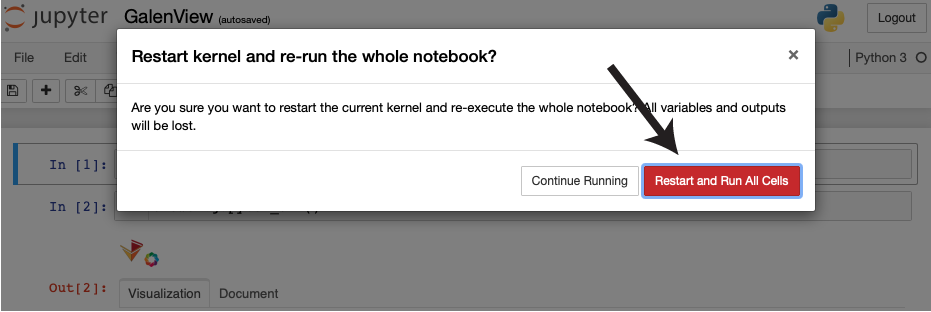
| A window will appear prompting you to restart the notebook. Click the red button. |
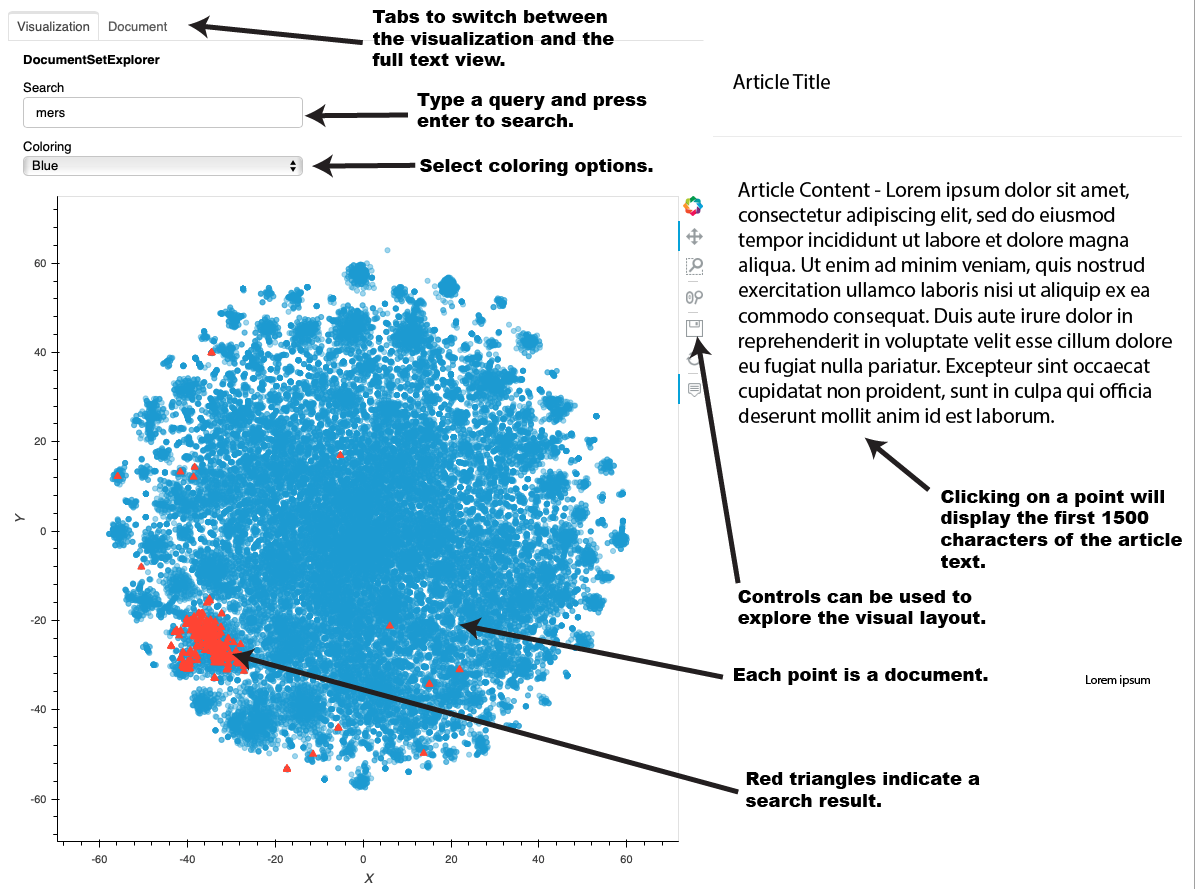
On the page, you will now see the full interface. Each point in the graph represents a document. Hovering your mouse over a point or group of points will display the title. Clicking on a point will cause the title and first 1500 characters (if full text is included in the CORD19 dataset) on the right hand side. Clicking on the "Document" tab will show the full text of the document.
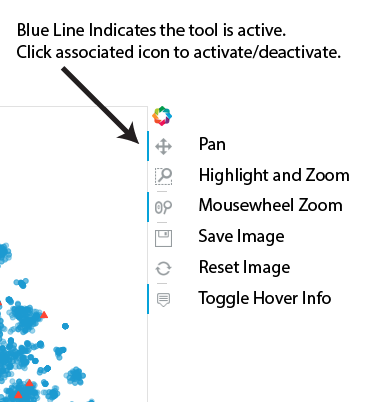 |
This interface lets you explore the various clusters of data by panning and zooming. The individual controls are described at the left. |
You can type in a query in the Search box and press enter. The points corresponding to the matching documents will turn into red triangles. Note that the documents are indexed using a python library called "whoosh" which contains a query language that lets you use advanced syntax such as boolean and proximity queries. More information on that query syntax can be found here.
This following is a short description of each part of the user interface:
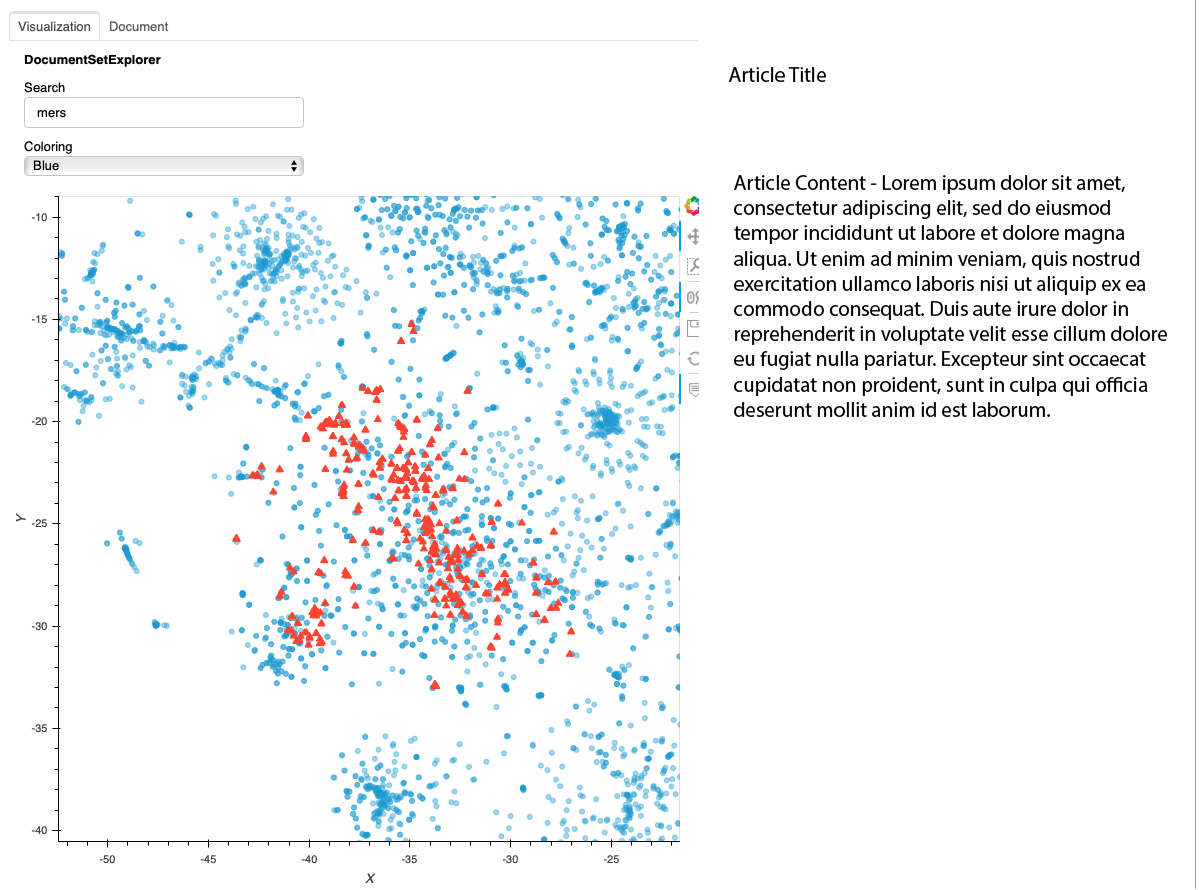
In the above example, the user has searched for the term "mers." The documents that reference "mers" are mostly found in a particular cluster at the lower left. Note that because of how the layout algorithm works, the cluster in your interface may be different. By using the controls, the user can zoom in on that cluster. The following image shows what that might look like. Note that the "mers" matches are in a cluster, but you can see similar clusters nearby. Through hovering, zomming, panning, and clicking, the user can explore the data set.
(Note: This section requres python development expertise)
In the paper associated with this codebase, compression-based analytics were used to color the documents according to how well they matched snippets of text provided by subject matter experts. The code to perform the compression-based analytics is not included in this codebase. However, it is possible to score documents in many different ways (even by something like year of publication). If you have a methodology for scoring the documents, you can get them into the visualization using the following steps:
-
Run the "Data Download and Configuration" step above. When it finishes, there will be a file called "metadata.df.pickle." in the data directory. This is a pandas DataFrame that can be read using pandas.read_pickle. The index (and integer) corresponds with the integer used by cotools in the cord-19-tools and is how the tool identifies the documents.
-
Create a score per document using whatever means is useful for your subject matter experts.
-
Append those scores as a new column to the DataFrame from step 1.
Write it back out to metadata.df.pickle. -
Create a Jupyter Notebook and create the visualization using the following code (where "myscore" is the name of column you added):
from sandia.galen.viewer import DocumentSetExplorer
display(HTML("<style>.container { width:100% !important; }</style>"))
hv.extension('bokeh')
d = DocumentSetExplorer(scoring_cols=["myscore"])
d.main_viz()If you want to test how this works before trying your own score, you can skip steps 2 and 3 and use "X" in place of "myscore." This will simply color the points according to their X value.
-
A large percentage of the documents do not have associated with them. All documents are arranged in the 2D space using both the title and content. But the search only searches the content unless you explicitly specify the title (using title:query). This is by design but can be confusing when navigating the space. This could be changed in a future version.
-
There currently is no way to set aside a document. This is entirely possible within the holoviews framework but is not yet impelemented.
-
It would be useful to include author, date published, journal, and other fields in the metadata. This information for many of the documents are in the metadata file and could be incorporated in the hover and visualization.
-
Links to the original documents that include tables and figures is important. For the documents that contain pubmed ids in the metadata, this is straight forward to do.
-
Implement better parsing of the documents, especially adding an ability to jump directly to different sections, depending on the specific question being asked. Sometimes, for example, it would be helpful to jump directly to the abstract. In other cases, jumping directly to the "methods" section would be best. Parsing research papers in this way is hard, but to the extent it can be done, the right hand summary pane could be customizable.
-
Add in paragraph spacing to increase readability. Right now, the document display used that cotools returns. But the underlying text is generally stored in a way that is better broken up and could be displayed with better paragraph breaks.