McCaD: Multi-Contrast MRI Conditioned, Adaptive Adversarial Diffusion Model for Accurate Healthy and Tumor Brain MRI Synthesis
This repo contains the official Pytorch implementation for McCaD.
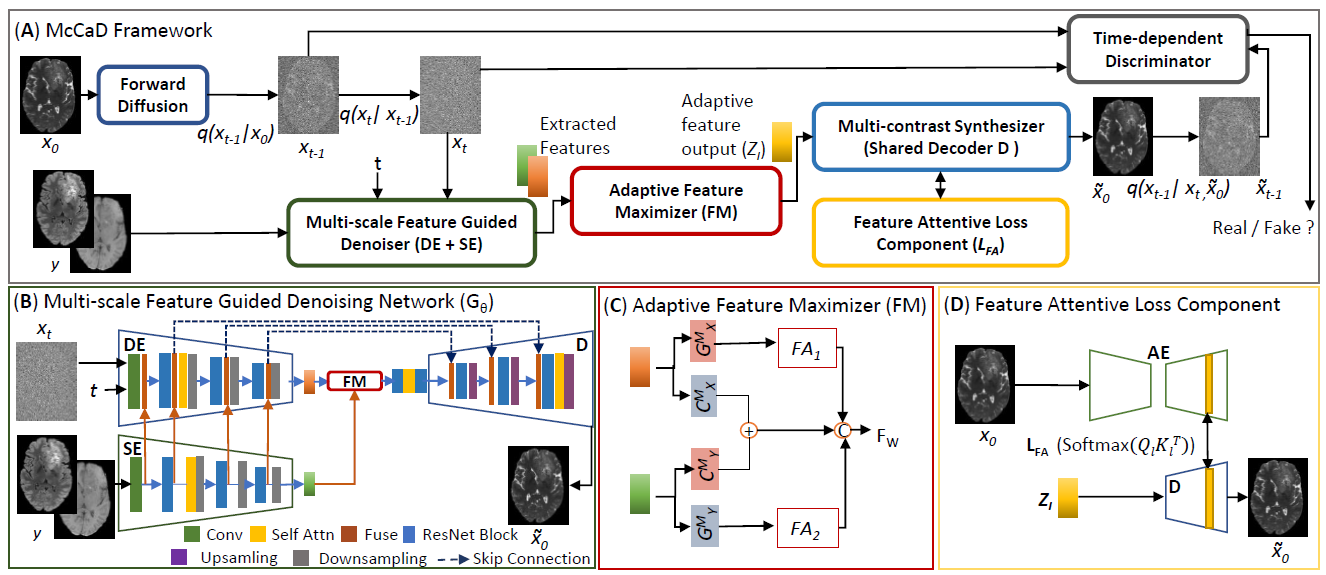
Network architecture of McCaD. A: Overall Architecture, B: Muti-scale Feature Guided Denosing Network, C: Adaptive Feature Maximizer, D: Feature Attentive Loss.
Environment
Please prepare an environment with python>=3.8, and then run the command "pip install -r requirements.txt" for the dependencies.
Data Preparation
For experiments, we used two datasets:
- Tumor Dataset : BRaTS Dataset
- Healthy Dataset
Dataset structure
data/
├── BRATS/
│ ├── train/
│ │ ├── T1.mat
│ │ └── T2.mat
│ │ └── FLAIR.mat
│ ├── test/
│ │ ├── T1.mat
│ │ └── T2.mat
│ │ └── FLAIR.mat
│ ├── val/
│ │ ├── T1.mat
│ │ └── T2.mat
│ │ └── FLAIR.mat
Use converter.py to convert 3D MRI data to 2D slices and save as .mat
Train Autoencoder
update input_path and output_path in train_autoencoder.py
python train_autoencoder.py --image_size 256 --exp exp_autoencoder --num_channels 1 --num_channels_dae 64 --ch_mult 1 1 2 2 4 4 --num_timesteps 4 --num_res_blocks 2 --batch_size 1 --num_epoch 40 --ngf 64 --embedding_type positional --ema_decay 0.999 --r1_gamma 1. --z_emb_dim 256 --lr_d 1e-4 --lr_g 1.6e-4 --lazy_reg 10 --num_process_per_node 1
Train McCaD
update input_path, output_path and the autoencoder checkpoint path in train_mccad.py
python train_mccad.py --image_size 256 --exp exp_brats_flair --num_channels 1 --num_channels_dae 64 --ch_mult 1 1 2 2 4 4 --num_timesteps 4 --num_res_blocks 2 --batch_size 1 --num_epoch 50 --ngf 64 --embedding_type positional --ema_decay 0.999 --r1_gamma 1. --z_emb_dim 256 --lr_d 1e-4 --lr_g 1.6e-4 --lazy_reg 10 --num_process_per_node 1
Test McCaD
update input_path and output_path in below cmd accordingly
python test_mccad.py --image_size 256 --exp exp_brats_flair --num_channels 1 --num_channels_dae 64 --ch_mult 1 1 2 2 4 4 --num_timesteps 4 --num_res_blocks 2 --batch_size 1 --embedding_type positional --z_emb_dim 256 --contrast1 T1 --contrast2 T2 --which_epoch 50 --gpu_chose 0 --input_path '/data/BRATS/' --output_path '/results'
Acknowledgements This repository makes liberal use of code from Tackling the Generative Learning Trilemma