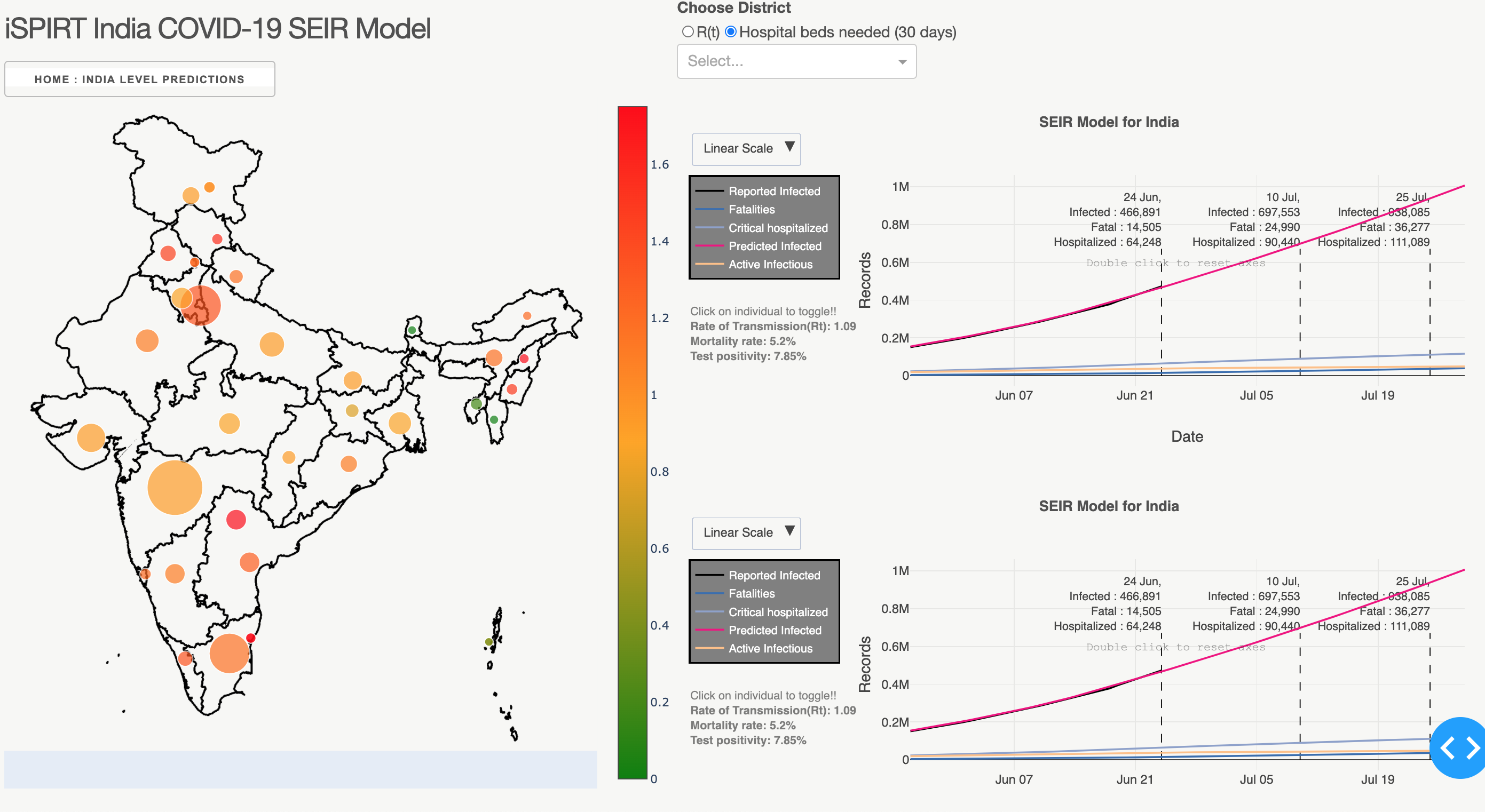
Implements a Network SEIR compartmental Model for forecasting COVID in India.
- Python 3
- AWS Account With S3 permission
Set following environment variables
ACCESS_KEY: "",
SECRET_KEY: "",
OPTIMIZER_ACCESS_KEY: "",
OPTIMIZER_SECRET_KEY: "",
open setup.py and add names of 2 buckets which will be used to store processed files and to create that run the script:
python script.py
Set custom configuration in config/resources.ini and should add same bucket names defined in above step.
Following dataset is used to build model:
https://api.covid19india.org/districts_daily.json
https://api.covid19india.org/csv/latest/statewise_tested_numbers_data.csv
https://api.covid19india.org/csv/latest/state_wise_daily.csv
https://api.covid19india.org/csv/latest/state_wise.csv
https://api.covid19india.org/csv/latest/district_wise.csv
build model locally by running below command:
python execute_seri_model.py
Once the model is successfully build it will upload data files to configured buckets.
We suggest you to create a virtual environment for running this app with Python 3. Clone this repository and open your terminal/command prompt in the root folder.
git@github.com:seirforindia/seirdistrictmodel.git
cd seirdistrictmodel
python3 -m virtualenv venv
In Unix system:
source venv/bin/activate
In Windows:
venv\Scripts\activate
Install all required packages by running:
pip install -r requirements.txt
Set following environment Variables:
Run this app locally by:
python app.py
Click on individual state from choropleth map to visualize state-specific flight delays in other plots and datatable, drag along time-series, click on single bar or drag along scatters to know flight details in the table.
Create Your Own Model SEIR Model
For Creating custom zone your must create global and nodal config with custom values. The global and nodal config has
{
"pop_frac": [0.44, 0.35, 0.15, 0.06],
"rate_frac": [1, 1, 1, 1],
"rates": 2.3,
"param": [
{
"intervention_date": "04-02-2020",
"rate_frac": [0.2, 0.2, 0.2, 0.2],
"intervention_type": "global"
}
],
"CFR": [0.01,0.065,0.125,0.25],
"P_SEVERE": [0.15, 0.35, 0.6, 0.9],
"I0": 50,
"E0": 75,
"D_death": 14,
"D_hospital_lag": 5,
"D_incubation": 5.2,
"D_infectious": 2.9,
"D_recovery_mild": 11.1,
"D_recovery_severe": 28.6,
"Fatal0": 0,
"Mild0": 0,
"R0": 0,
"R_Fatal0": 0,
"R_Mild0": 0,
"R_Severe0": 0,
"S0": -1,
"Severe0": 0,
"Severe_H0": 0
}{
"node": "Delhi Cantonment",
"pop": 116352,
"nodal_param_change": [
{
"intervention_date": "04-03-2020",
"intervention_type": "local",
"delI": 20,
"rate_frac": [0.4, 0.4, 0.4, 0.4]
}
]
}pop_frac: population fractions of different age groups: [0-19,20-39,40-59,60+]
rate_frac: rate multiplier of different age groups (default value of 1 corresponds to a rate of ~2.3)
rates: basic reproduction number R0 is the number of secondary infections each infected individual produces (largest eigenvalue chosen from the 4×4 matrix used to model the four age categories)
param: global intervention is a time event that impacts the parameters of the simulation (for eg: lockdown)
intervention_date: date when intervention was put in place
intervention_type: distinguishes between global and nodal interventions
intervention_day: number of days after which the intervention was put in place (void from latest version)
CFR: case fatality rate of different age groups (percentage of infected people who pass away)
P_SEVERE: hospitalization rate of different age groups (percentage of infected people who need to be hospitalized)
I0: initial number of infectious persons at the start of the simulation
E0: initial number of exposed persons at the start of the simulation
D_death: average time duration an individual takes to move from infection to death
D_hospital_lag: average time duration for an infected person to be hospitalised (assuming the individual is hospitalised)
D_incubation: time duration an exposed person incubates for before getting infected
D_infectious: time duration an infectious individual (symptomatic and asymptomatic) is shedding the virus
D_recovery_mild: recovery time for mild cases (non-hospitalised individuals)
D_recovery_severe: average length of stay in a hospital for severe cases
Fatal0: initial number of fatal persons at the start of the simulation
Mild0: initial number of recovering mild cases at the start of the simulation
R0: initial number of removed persons (recovered or dead) at the start of the simulation
R_Fatal0: initial number of removed fatal cases at the start of the simulation
R_Mild0: initial number of removed mild cases at the start of the simulation
R_Severe0: initial number of removed hospitalized cases at the start of the simulation
S0: initial number of susceptible persons (number of people who are not immune to the disease at the start of the simulation, typically encompasses the entire population)
Severe0: initial number of severe cases recovering at home at the start of the simulation
node: name of the node ie the geographical boundary for which the simulation is executed
t0: offset value ie the starting day for the specific simulation in comparison to the simulation’s reference
pop: total population within the node’s geographic boundary
nodal_param_change: node-wise interventions ie time events that impacts only the geographical area for which the node is defined
intervention_date: date when intervention was put in place
intervention_type: distinguishes between global and nodal interventions
intervention_day: number of days after which the intervention was put in place
rate_frac: rate multiplier of different age groups (default value of 1 corresponds to a rate of ~2.3)