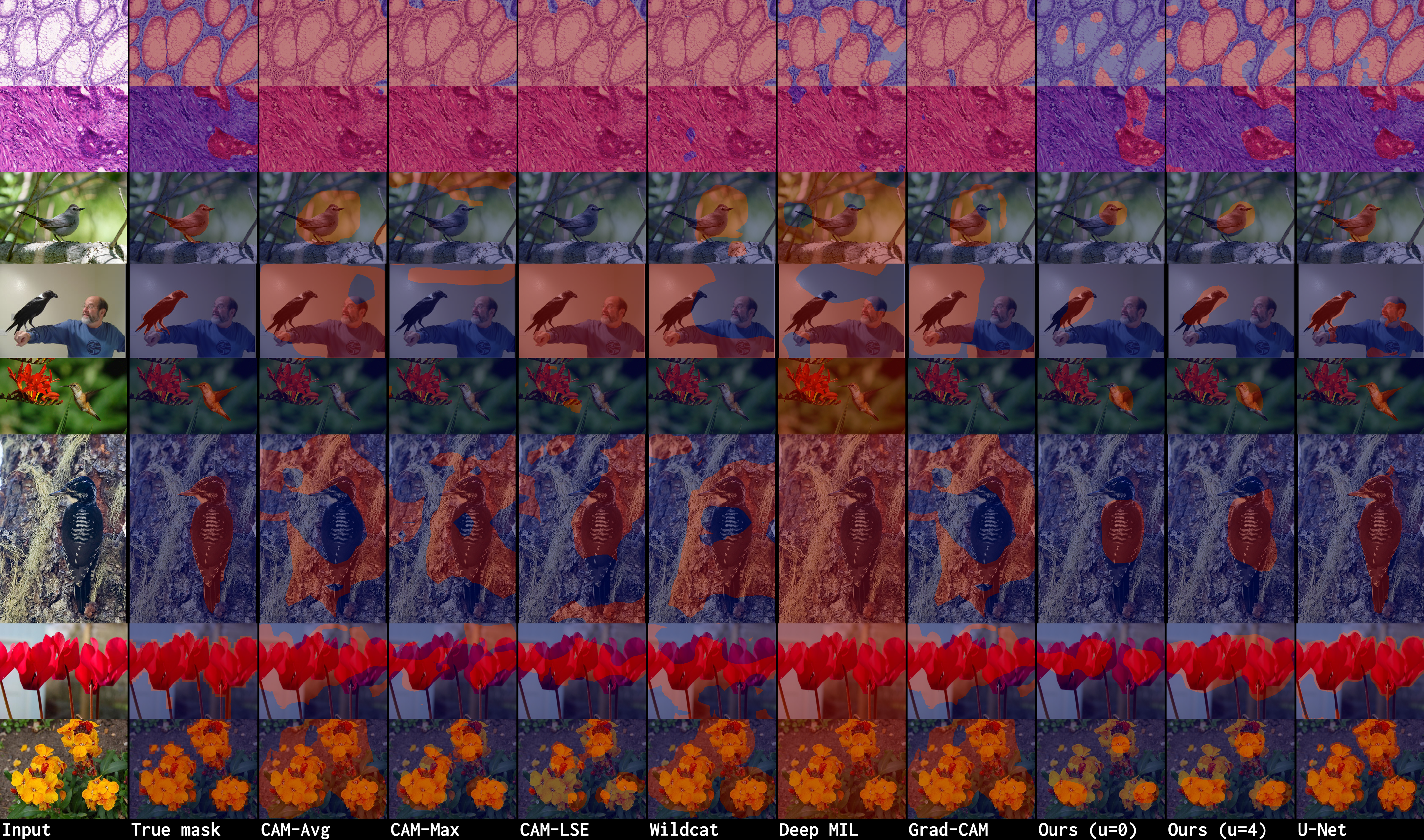
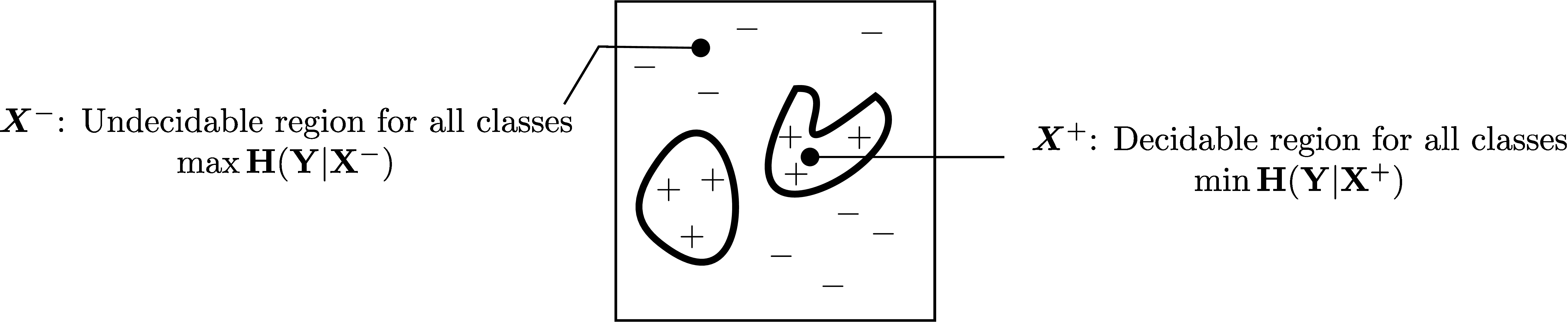
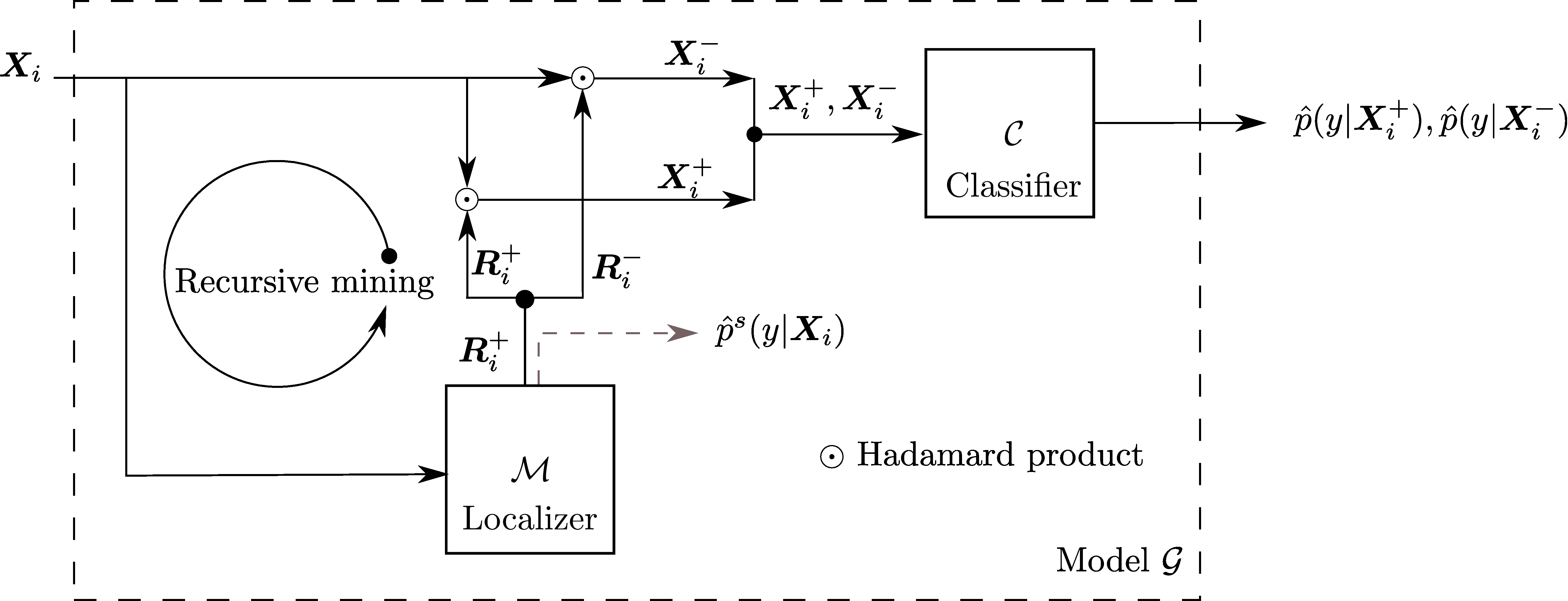
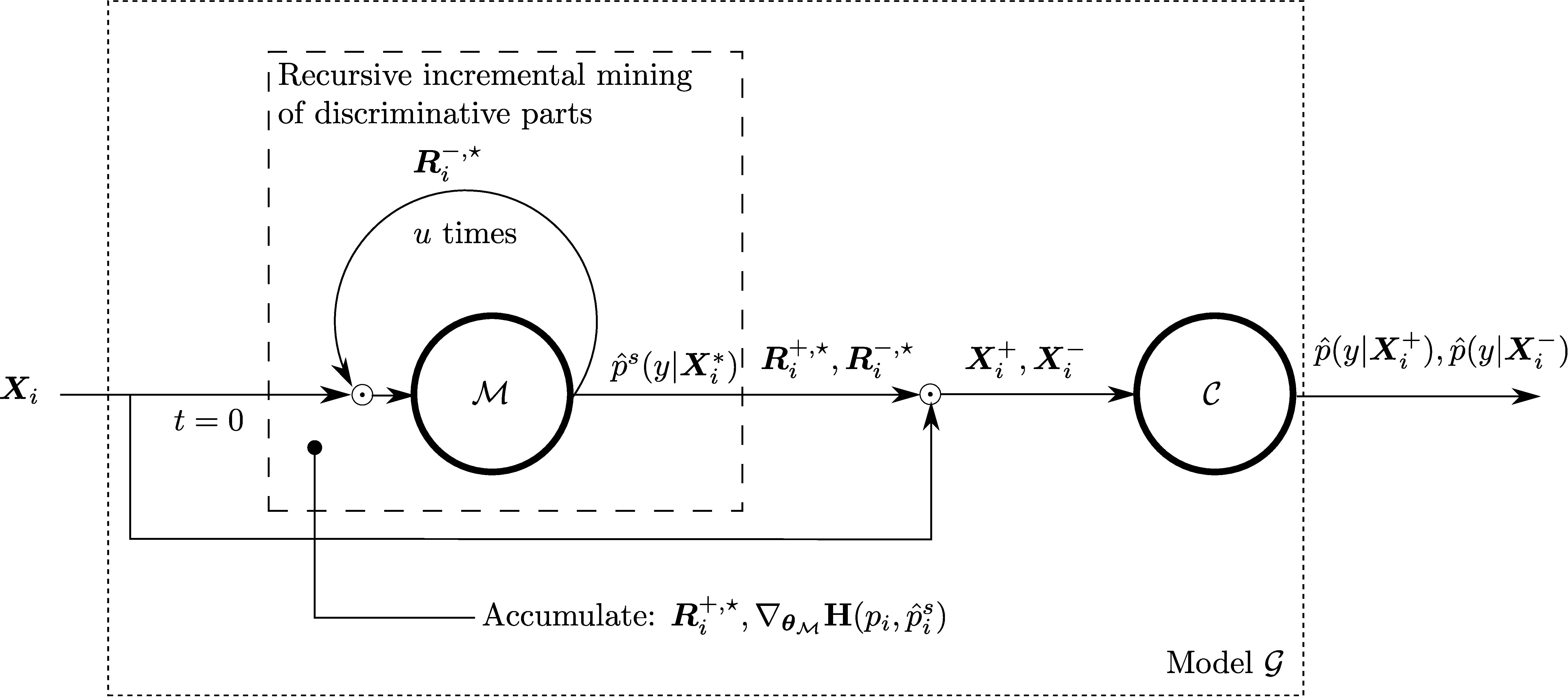
Pytorch code for: Min-max Entropy for Weakly Supervised Pointwise Localization (arxiv.org/abs/1907.12934)
- If you use this code, please cite our work:
@article{belharbi2019weakly,
title={Min-max Entropy for Weakly Supervised Pointwise Localization},
author={Belharbi, S. and Rony, J. and Dolz, J. and Ben Ayed, I. and McCaffrey, L. and Granger, E.},
journal={CoRR},
volume={abs/1907.12934},
year={2019}
}
- Demo
- Method overview
- Datasets
- Requirements
- How to run the code
- Reproducibility
- MultiGPU support and reproducibility
- Synchronized-Batch-Norm support (MultiGPUs)
1. Prediction (test samples. More results with high resolution are in ./demo.md):
- GlaS: ./download-glas-dataset.sh.
- Caltech-UCSD Birds-200-2011: ./download-caltech-ucsd-birds-200-2011-dataset.sh
- Oxford flower 102: ./download-Oxford-flowers-102-dataset.sh
You find the splits in ./folds. The code that generated the splits is ./create_folds.py.
We use torch_nightly-1.2.0.dev20190616 and Python 3.7.0. For installation, see ./dependencies for a way on how to install the requirements within a virtual environment.
Installing some packages manually:
- Pytorch:
pip install https://download.pytorch.org/whl/nightly/cu100/torch_nightly-1.2.0.dev20190616-cp37-cp37m-linux_x86_64.whl - Torchvision:
pip install torchvision==0.2.2.post3 - SPAMS:
wget http://spams-devel.gforge.inria.fr/hitcounter2.php?file=file/37660/spams-python.anaconda-v2.6.1-2018-08-06.tar.gz$ tar -xf 'hitcounter2.php?file=file%2F37660%2Fspams-python.anaconda-v2.6.1-2018-08-06.tar.gz'$ cd spams-2.6.1/$ python setup.py install
- PyDenseCRF for CRF post-processing (commit:
4d5343c398d75d7ebae34f51a47769084ba3a613):pip install git+https://github.com/lucasb-eyer/pydensecrf.git@4d5343c398d75d7ebae34f51a47769084ba3a613
python train_deepmil.py --cudaid your_cuda_id --yaml basename_your_yaml_fileYou can override the values of the yaml file using command line:
python train_deepmil.py --cudaid 1 --yaml glas-no-erase.yaml --kmin 0.09 --kmax 0.09 --dout 0.0 --epoch 2 \
--nbrerase 0 --epocherase 1 --stepsize 40 --bsize 8 --modalities 5 --lr 0.001 --fold 0 --wdecay 1e-05 --alpha 0.2See all the keys that you can override using the command line in tools.get_yaml_args().
- All the experiments, splits generation were achieved using seed 0. See ./create_folds.py
- All the results in the paper were obtained using one GPU.
- Please report any issue with reproducibility.
We hard-coded some paths (to the data location). For anonymization reasons, we replaced them with fictive paths. So, they won't work for you. A warning will be raised with an indication to the issue. Then, the code exits. Something like this:
warnings.warn("You are accessing an anonymized part of the code. We are going to exit. Come here and fix this "
"according to your setup. Issue: absolute path to Caltech-UCSD-Birds-200-2011 dataset.")The yaml files in ./config_yaml are used for each dataset.
We use the implementation in (https://github.com/lucasb-eyer/pydensecrf).
To be able to use it, first install the package. Then, you can use the class: tools.CRF(). Read its documentation for more details. The parameters of the CRF are in ./crf_params.py. For CUB5, CUB, OxF, we used crf_params.cub.
We took a particular care to the reproducibility of the code.
- The code is reproducible under Pytorch reproducibility terms.
Completely reproducible results are not guaranteed across PyTorch releases, individual commits or different platforms. Furthermore, results need not be reproducible between CPU and GPU executions, even when using identical seeds.
- The code is guaranteed to be reproducible over the same device INDEPENDENTLY OF THE NUMBER OF WORKERS (>= 0). You have to use a seed in order to obtain the same results over two identical runs. See ./reproducibility.py
- Samples can be preloaded in memory to avoid disc access. See ./loader.PhotoDataset(). DEFAULT SEED IS 0 which we used in all our experiments.
- Samples can be preloaded AND preprocessed AND saved in memory (for inference, i.e., test). However, for large dataset, and when the pre-processing aims at increasing the size of the data (for instance, upsampling), this is to be avoided. See ./loader.PhotoDataset()
- We decorated sensitive operations that use random generators by a fixed seed. This allowed more flexibility in terms of reproducibility. For instance, if you decide to switch off pre-processing samples of a dataset (given that such operation relies heavily on random generators) and do the processing on the fly, such a decorator seed allows the state of the random generators in the following code to be independent of the results of this switch. This is a work-around. In the future, we consider more clean, and easy way to make the operations that depend on random generator independent of each other.
Due to some issues in the prng state of Pytorch 1.0.0, we moved to Nightly build version 1.2.0.dev20190616
(https://download.pytorch.org/whl/nightly/cu100/torch_nightly-1.2.0.dev20190616-cp37-cp37m-linux_x86_64.whl) which
seems to have fixed a huge glitch in the rng. (merge). Please refer
to this post.
We use this version to make multigpu case reproducible (best).
7. MultiGPU support and reproducibility (100% reproducibility not guaranteed. Sometimes the results are different, but most of the time the results are constant):
- The code supports MultGPUs.
- Despite our effort to make the code reproducible in the case of multigpu, we achieve reproducibility but, sometimes it breaks. See this.
The code supports synchronized BN. By default, SyncBN is allowed as long as multigpu mode is on. You can prevent using SynchBN, and get back to the standard Pytroch non-synchronized BN, using bash command, before running the code:
# $ export ACTIVATE_SYNC_BN="True" ----> Activate the SynchBN
# $ export ACTIVATE_SYNC_BN="False" ----> Deactivate the SynchBNAll the credits of the SynchBN go to Tamaki Kojima(tamakoji@gmail.com) (https://github.com/tamakoji/pytorch-syncbn).