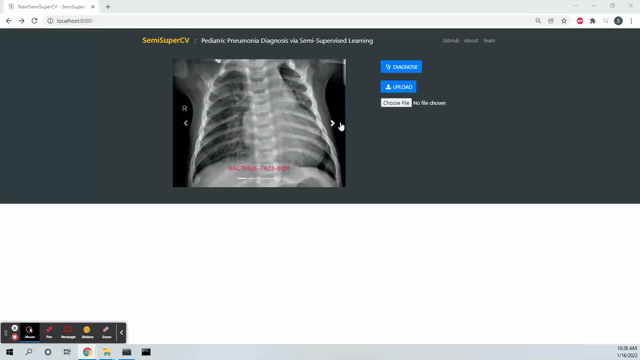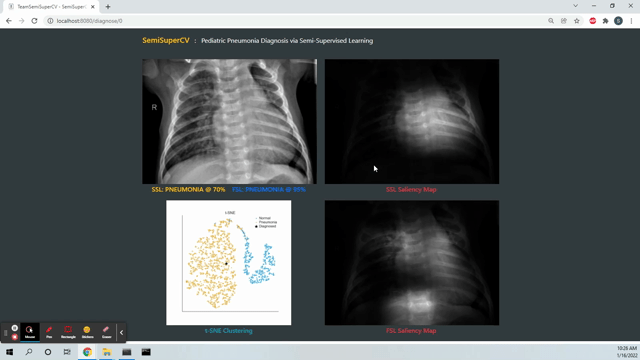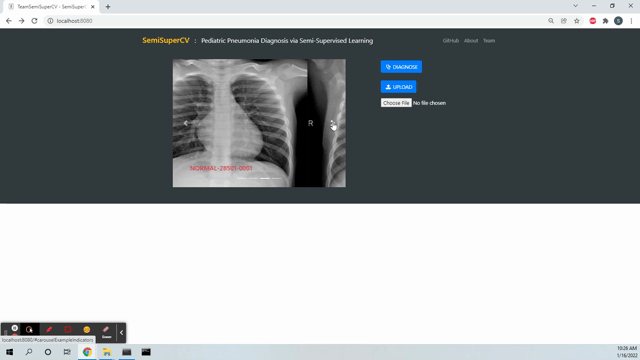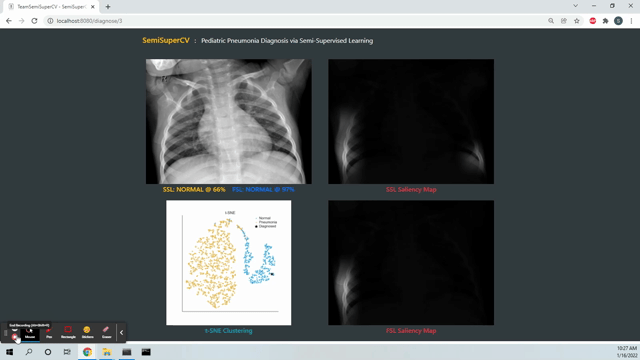
Live Deployment! The deployed SemiSuperCV app predicts if a patient has pneumonia based on a chest X-ray. You can try the app at https://webapp-mcdo4dmd2a-uc.a.run.app. The first couple of runs might have high latency due to cold start times on Google Cloud Run.
Fully supervised approaches need large, densely annotated datasets. Only hospitals that can afford to collect large annotated datasets can utilize these approaches to aid their physicians. The project goal is to utilize self-supervised and semi-supervised learning approaches to significantly reduce the need for fully labeled data. In this repo, you will find the project source code, training notebooks, and the final TensorFlow 2 saved model used to develop the web application for detecting pediatric pneumonia from chest X-rays.
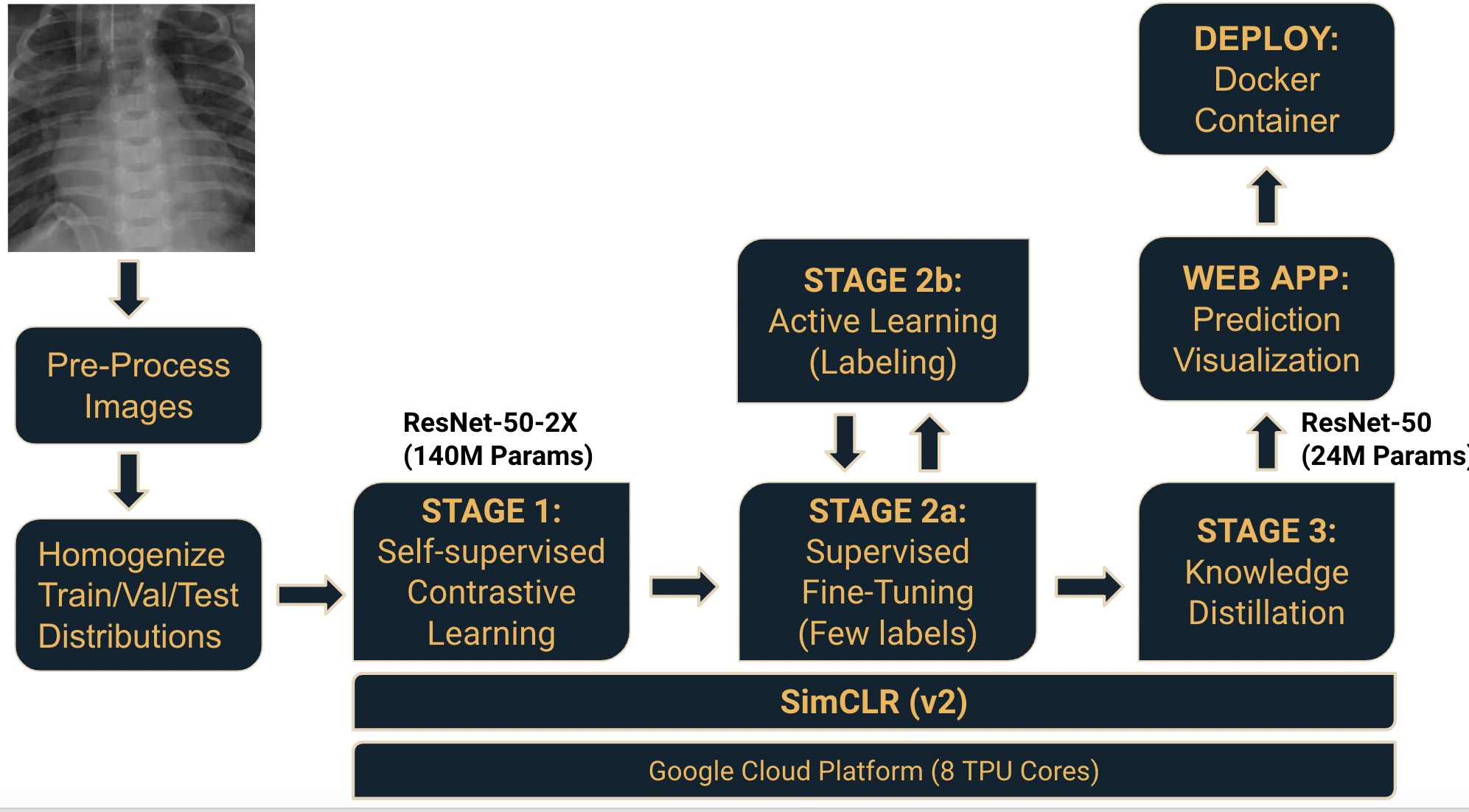
The semi-/self-supervised learning framework used in the project comprises three stages:
- Self-supervised pretraining
- Supervised fine-tuning
- Knowledge distillation using unlabeled data
Refer to Google research team's paper “SimCLRv2 - Big Self-Supervised Models are Strong Semi-Supervised Learners” for more details regarding the framework used.
The training notebooks for Stage 1, 2, and 3 can be found in the notebooks folder. The project report and the final presentation slides can be found in the docs folder.
Resources
Follow the instructions below to run the app locally with Docker.
Once you have Docker installed, clone this repo
git clone https://github.com/TeamSemiSuperCV/semi-superNavigate to the webapp directory of the repo.
cd semi-super/webappNow build the container image using the docker build command. It will take few minutes.
docker build -t semi-super .Start your container using the docker run command and specify the name of the image we just created:
docker run -dp 8080:8080 semi-superAfter a few seconds, open your web browser to http://localhost:8080. You should see our app.
| Labels | Stage 1 (self-supervised) |
|---|---|
| No labels used | 99.99% |
| Labels | FSL1 | Stage 2 (finetuning) | Stage 3 (distillation) |
|---|---|---|---|
| 1% | 85.2% | 94.5% | 96.3% |
| 2% | 87.2% | 96.8% | 97.6% |
| 5% | 86.0% | 97.1% | 98.1% |
| 100% | 98.9% |
Despite using only a fraction of labels, our Stage 2 and Stage 3 models achieved test accuracies comparable to a 100 percent fully supervised (FSL) model. Refer to the project report and the final presentation slides in the docs folder for a more detailed discussion and findings.
We have taken the SimCLR framework code from Google Research and heavily modified it for the purpose of this project. We have added a knowledge distillation feature along with several other changes. Following these changes and improvements, knowledge distillation can be performed on Google Cloud TPU cluster, which reduces training time significantly.
Footnotes
-
Fully-supervised model performance for benchmarking purposes. ↩