This repository holds all the recipes used to run the Poseidon Framework's Minotaur Workflow and create poseidon packages.
The Minotaur workflow takes in sequencing metadata and URLs of publicly archived
raw sequencing data (provided by the community) and processes them in a flexibly
configurable yet reproducible manner to produce poseidon packages. These
poseidon packages are then added to the Poseidon Minotaur Archive (PMA), and
made available to everyone through
trident, the poseidon
server-client infrastructure.
The Minotaur Workflow consists of three parts:
- Creating the Poseidon Package recipe
- Processing of the public data with nf-core/eager
- Poseidon Package preparation for upload to the PMA
Details on each part can be found below.
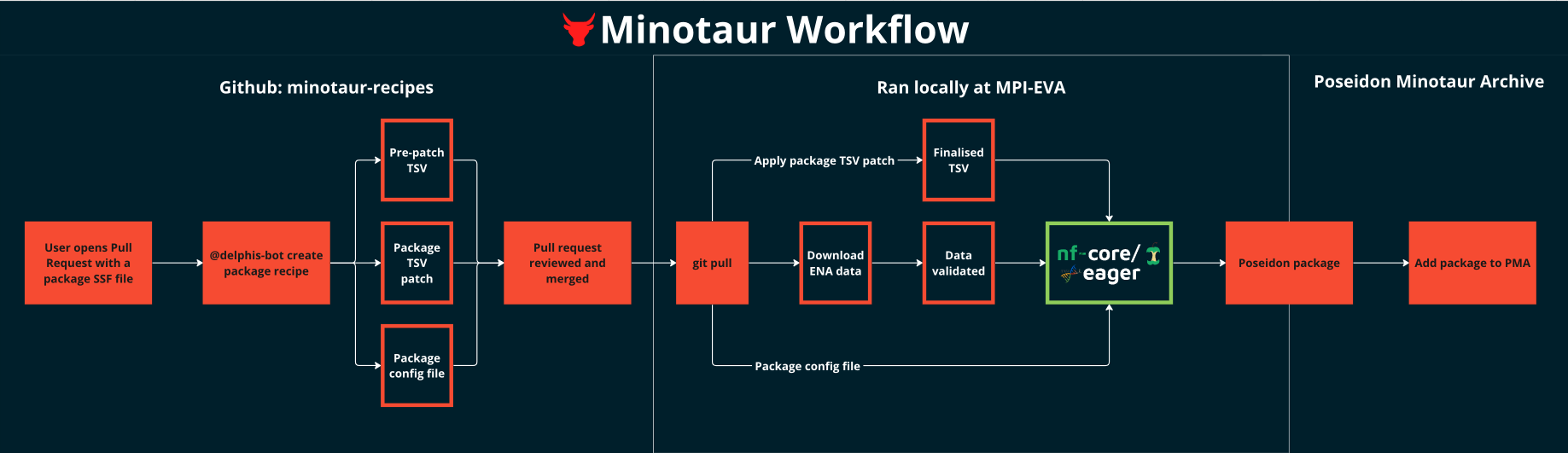
This is the community-facing entrypoint to the workflow, and takes place on the
minotaur-recipes GitHub repository,
when a contributor has opened a pull request to add/update a package. Once the
required SSF file has been updated, delphis-bot will create all the
SSF-associated auxilliary files required for processing.
Upon activation, delphis-bot will create:
- a precursor nf-core/eager TSV input file,
- the package
.configfile, - the package
tsv_patch.shthat can be ued to localise the TSV precursor into a valid input for nf-core/eager, - a
script_versions.txtfile, with the versions of the scripts used during the package recipe creation.
For a step-by-step guide on how to contribute to the PMA, see this guide.
This step takes place locally at MPI-EVA. The machinery described in the poseidon-eager GitHub repository uses the package recipe to:
- download the raw data from the public archive URLs in the SSF file (using
scripts/download_ena_data.pyandscripts/run_download.sh) - Validate the downloaded data, and create symlinks with clearer naming
(
scripts/validate_downloaded_data.sh). This allows the one-to-many relationship between raw data andposeidon_ids. - Apply the
*_tsv_patch.shof the package recipe to create the finalised nf-core/eager TSV. - Use
run_eager.shto run nf-core/eager.- This uses the finalised TSV as its input
- And load the
.configof the package recipe to apply all default parameters, as well as any relevantCaptureTypeand package-specific parameters.
Once the data is processed, the genotyping output of nf-core/eager is used to create a poseidon package. The metadata included in the janno file for this package is then filled using descriptive statistics generated by nf-core/eager and information from the SSF file.