This project aims to be a comprehensive tool for building single cell models using Arbor. It strives to be self-contained, fast, and easy to use.
- Design morphologically detailled cells for simulation in Arbor.
- Load morphologies from SWC
.swc, NeuroML.nml, NeuroLucida.asc. - Define and highlight Arbor regions and locsets.
- Paint ion dynamics and bio-physical properties onto morphologies.
- Place spike detectors and probes.
- Export cable cells to Arbor’s internal format (ACC) for direct simulation.
- Import cable cells in ACC format
This project is under active development and welcomes early feedback. Currently, Arbor master as of March 2021 is supported and bundled with the project. Note that the screenshots below are updated less frequently than the actual project. We aim for a formal release with Arbor 0.6 at which point the project will stabilise and receive regular updates alongside Arbor.
We welcome bug reports and feature requests, please use the issue tracker here on GitHub for these purposes. Building network simulation is out of scope for this project (we might offer a different tool, though).
Note The screenshots below are somewhat outdated, the current status offers quite a bit more.
- Rendering of cable cell as seen by Arbor.
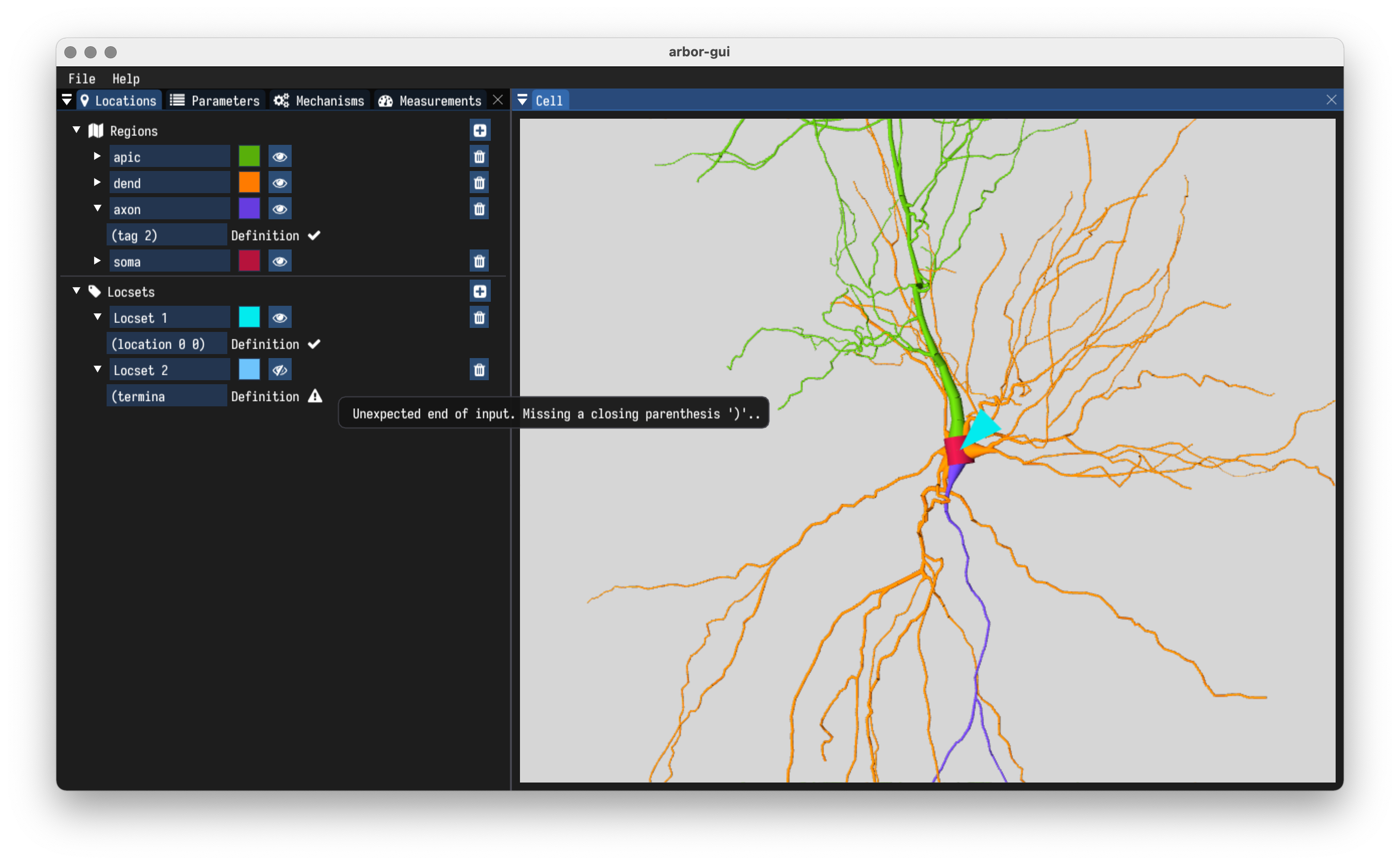
- Define locations in Arbor’s Locset/Region DSL.
- Live feedback by Arbor’s parser.
- Well-formed expressions are rendered immediately.
- Navigate with
- pan: arrow keys or C-drag,
- zoom: +/- or mouse wheel,
- rotate: mouse drag.
- Right-click to
- reset camera,
- snap-to a defined locset,
- set the background colour,
- tweak morphology orientation,
- toggle orientation guide,
- save the currently rendered image to disk.
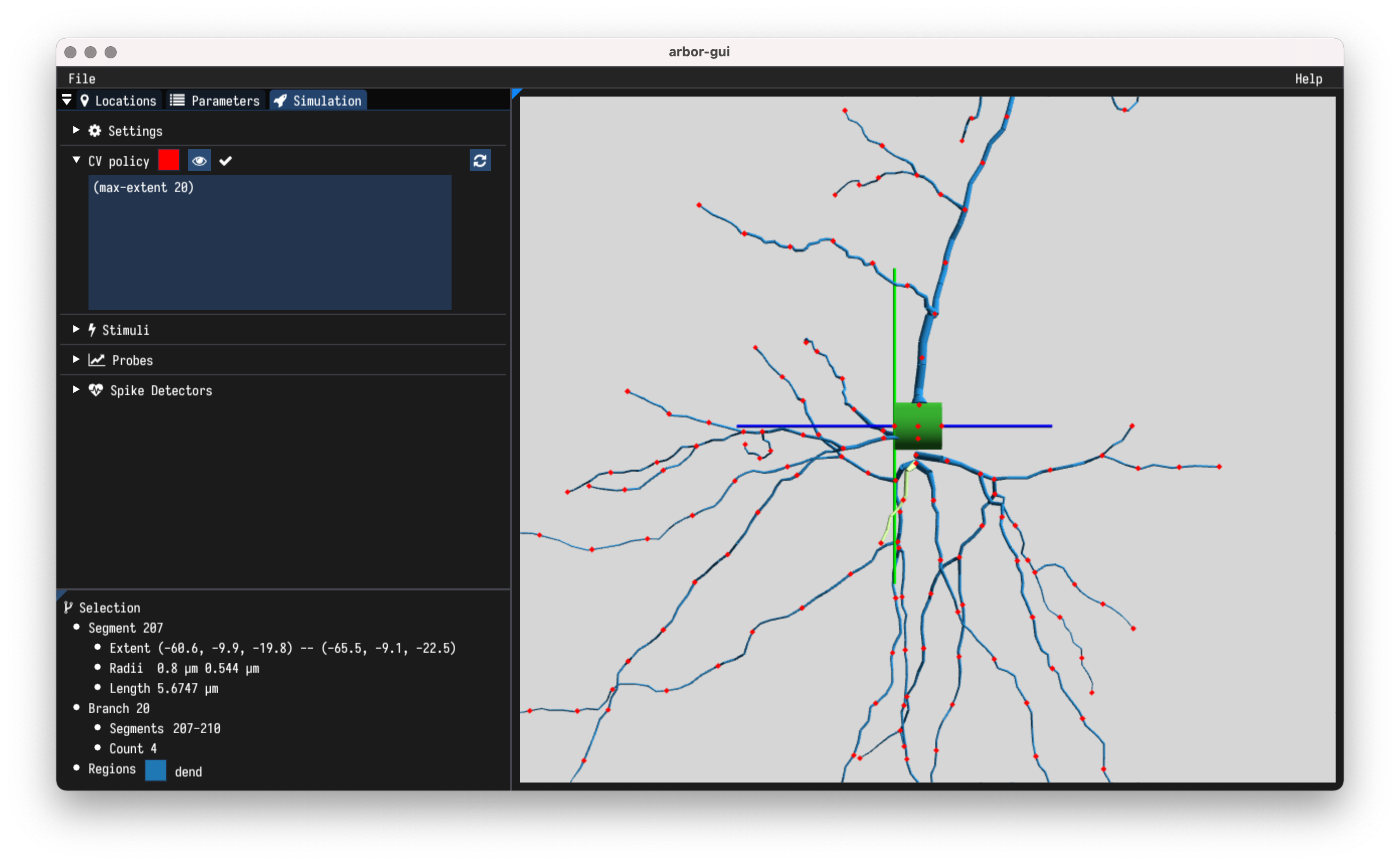
- Hover a segment to show
- containing branch and regions,
- geometry information.
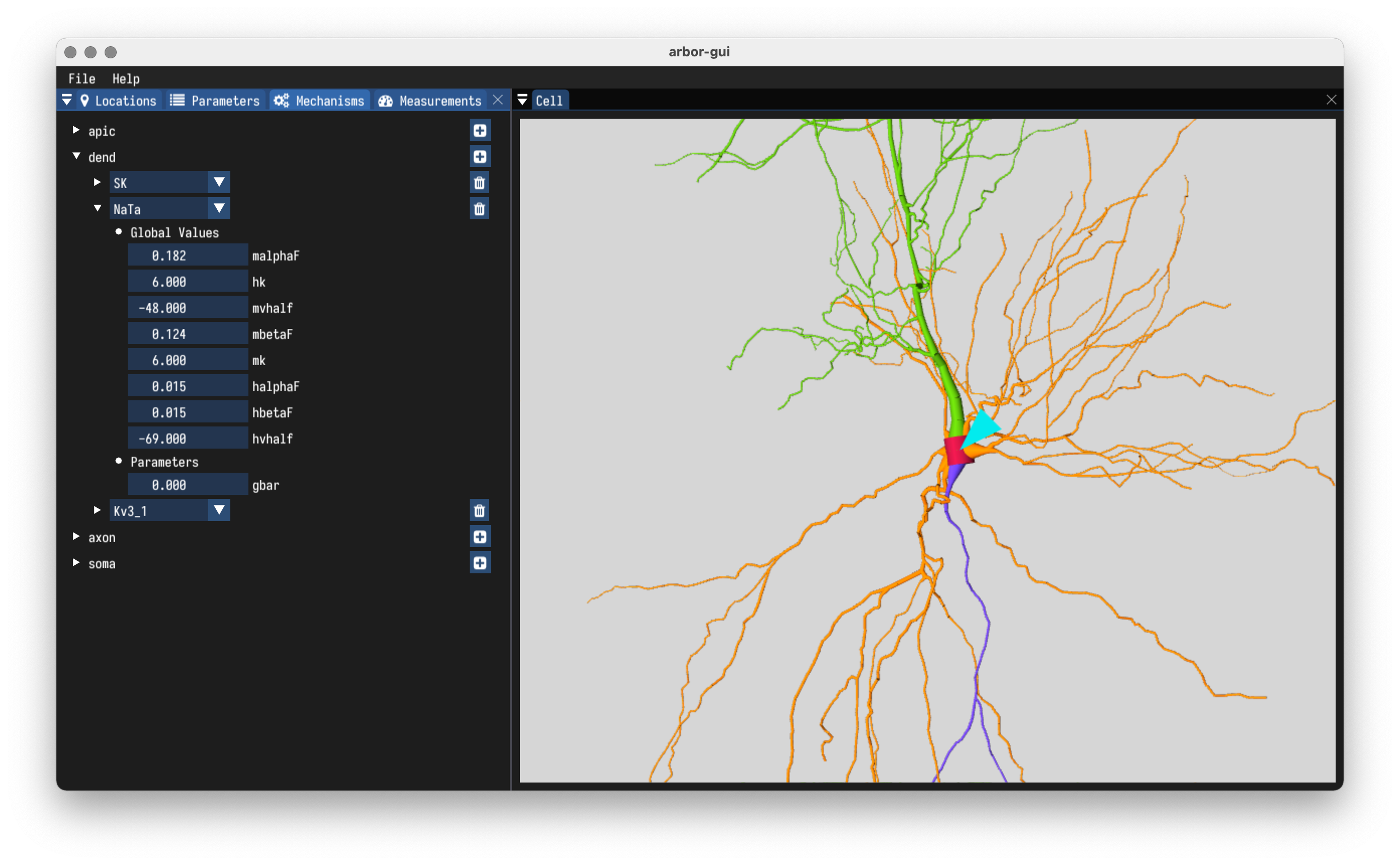
- Load mechanisms from built-in catalogues.
- Define ion species.
- Set parameters of mechanisms and ions.
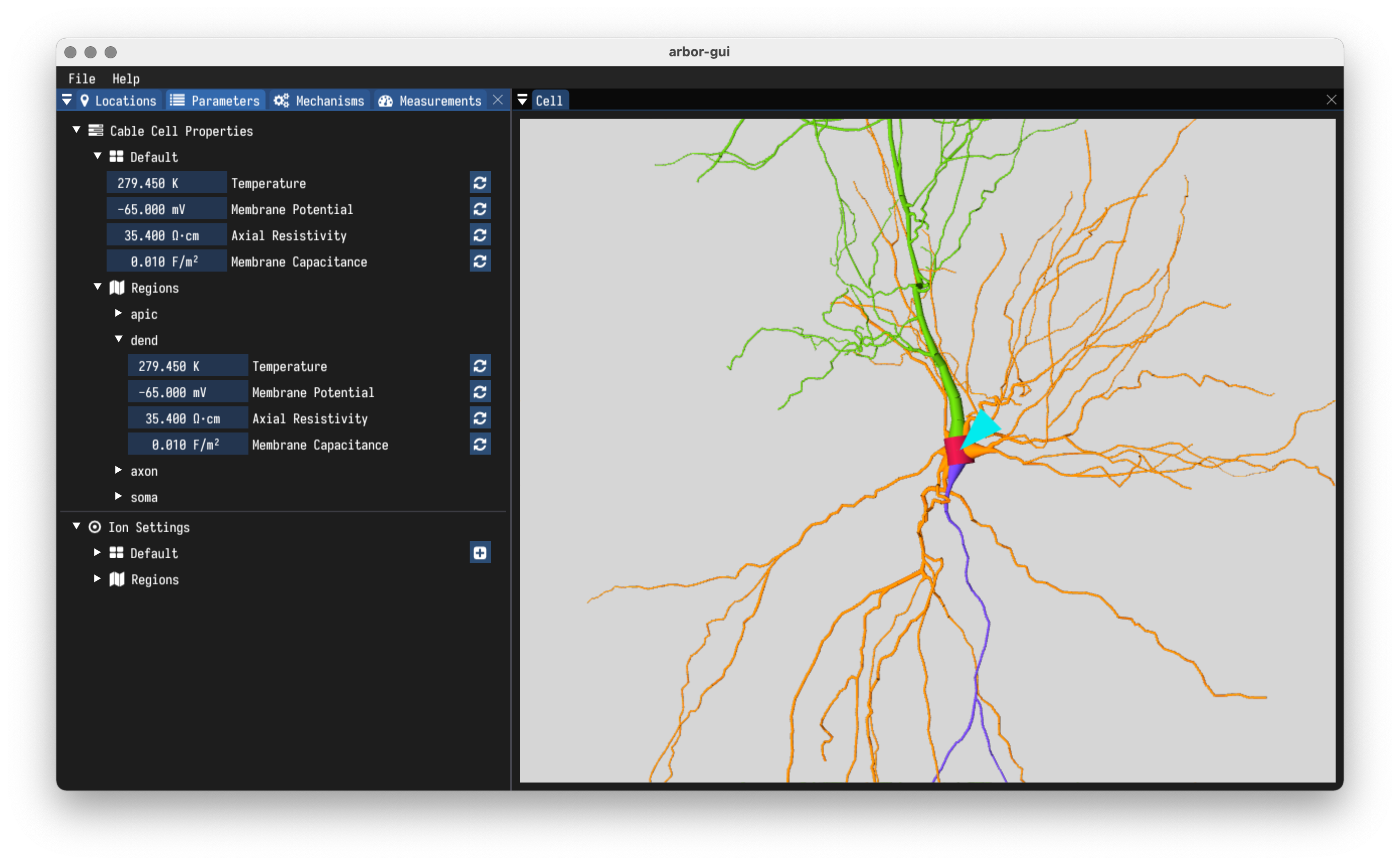
- Set global and cell level defaults.
- Set per-region parameters like temperature, resisitivities, and more.
- Set global and cell level defaults.
- Timestep and simulation interval.
- Add Probes, Spike Detectors.
- Set and visualise discretisation policy
- You can adjust the GUI layout by dragging and dropping windows and tabs.
- Dragging regions will change rendering order, so overlapping regions might be better visible.
Please install a OpenGL package confirming to your choice and platform. You will also need a recent (as in C++20 supported) C++ compiler.
Then, clone this repository like this
git clone --recursive https://github.com/thorstenhater/arbor-gui.git
this will bring in all the requirements apart from OpenGL.
Next, configure and build the project
cd arbor-gui mkdir build cd build cmake .. make install
Please use a recent version of Clang, as installed by brew for example. The project has been confirmed to build and run with Clang 11 on BigSur and Catalina using this line
cmake .. -DCMAKE_CXX_COMPILER=/usr/local/opt/llvm/bin/clang++ -DCMAKE_C_COMPILER=/usr/local/opt/llvm/bin/clang -DCMAKE_BUILD_TYPE=release
This project uses various open source projects, licensed under permissive open source licenses. See the respective projects for license and copyright details.
- Arbor: https://github.com/arbor-sim/arbor
- GLM for OpenGL maths: https://github.com/g-truc/glm
- GLFW for setting up windows: https://github.com/glfw/glfw
- Dear ImGUI library https://github.com/ocornut/imgui
- Iosevka font https://github.com/be5invis/Iosevka
- ForkAwesome icon set https://github.com/ForkAwesome/Fork-Awesome
- C++ icon bindings https://github.com/juliettef/IconFontCppHeaders
- fmt formatting https://github.com/fmtlib/fmt
- spdlog logger https://github.com/gabime/spdlog
- stb image loader https://github.com/nothings/stb
- Tracy profiler https://github.com/wolfpld/tracy.git
Test and example datasets include:
- A morphology model
dend-C060114A2_axon-C060114A5.asccopyright of the BBP, licensed under the CC BY-NC-SA 4.0 license.