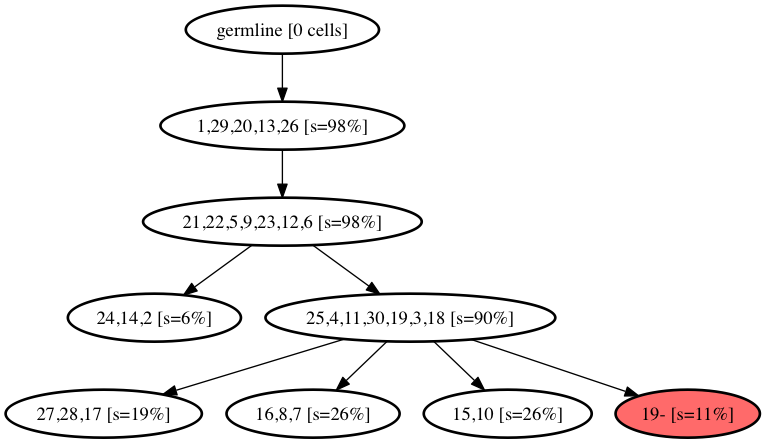
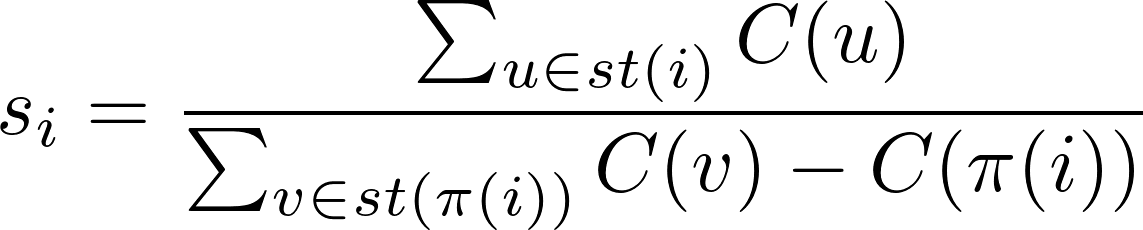reSASC is a new model and a robust framework based on Simulated Annealing for the inference of cancer progression from the SCS data. The main objective is to overcome the limitations of the Infinite Sites Assumption by introducing a novel phylogeny model DCS(k,j) that is a combination of the Dollo-k and Camin-Sokal-j that allows both the deletion and recurrences of mutations from the evolutionary history of the tumor.
reSASC can be downloaded and compiled easily using the following commands:
git clone git@github.com:sciccolella/reSASC.git
cd reSASC
makeSingle Cell file
The input file (specified by the -i parameter) is expected to be a ternary matrix file where the rows represent the cells and the columns the mutations. Each cell must be separated by a space or by a tab (\t). Each cell of the matrix can be:
| Value of cell | Meaning |
|---|---|
| I[i,j] = 0 | Mutation j is not observed in cell i |
| I[i,j] = 1 | Mutation j is observed in cell i |
| I[i,j] = 2 | There is no information for mutation j in cell i, i.e. low coverage |
An example of the input file can be seen in MGH36_scs.txt.
Mutations file
This optional file specifies the name of the mutations (parameter -e). Each mutation's name must be on a different line (separated by \n), and the names are assigned to columns from left to right in the input file. If this file is not provided, mutations are progressively named from 1 to the total number of mutations.
An example of the mutations' name file can be seen in MGH36_snv-names.txt.
Cells file
This optional file specifies the name of the cells (parameter -E). Each cell's name must be on a different line (separated by \n), and the names are assigned to rows from top to bottom in the input file. If this file is not provided, cells are progressively named from 1 to the total number of cells.
An example of the mutations' name file can be seen in MGH36_cell-names.txt.
FN rates file
This optional file specifies the false negative rate of the mutations (parameter -a). Each mutation's FN rate must be on a different line (separated by \n), and the rates are assigned to columns from left to right in the input file. If this file is not provided, it is necessary to select a single float value that will be interpreted as the FN rate for each mutation.
An example of the mutations' name file can be seen in MGH36_fn-rates.txt.
A single FN rate can be specified directly when running the program, e.g. -a 0.3.
Prior losses file
This optional file specifies the prior loss probability of the mutations (parameter -g). Each mutation's prior must be on a different line (separated by \n), and the probabilities are assigned to columns from left to right in the input file. If this file is not provided, it is possible to select a single float value that will be interpreted as the prior loss probability for each mutation. If the value is not provided it will be set to 1 by default.
An example of the mutations' name file can be seen in gammas.txt.
A single FN rate can be specified directly when running the program, e.g. -g 0.01.
Prior recurrent file
This optional file specifies the prior recurrent probability of the mutations (parameter -q). Each mutation's prior must be on a different line (separated by \n), and the probabilities are assigned to columns from left to right in the input file. If this file is not provided, it is possible to select a single float value that will be interpreted as the prior recurrent probability for each mutation. If the value is not provided it will be set to 1 by default.
An example of the mutations' name file can be seen in deltas.txt.
A single FN rate can be specified directly when running the program, e.g. -q 0.01.
Input Parameters (required)
-n [INT]: Number of cells in the input file.-m [INT]: Number of mutations in the input file.-k [INT]: k value of Dollo-k model.-j [INT]: j value of Camin-Sokal-j model.-a [FLOAT/STRING]: False Negative rate in the input file or path of the file containing different FN rates for each mutations.-b [FLOAT]: False Positive rate in the input file.-i [STRING]: Path of the input file.
Model parameters (optional)
-d [INT]: Maximum number of total deletions allowed in the solution. By default the value is set to have no restriction (+INF).-c [INT]: Maximum number of total recurrences allowed in the solution. BY default the value is set to have no restriction (+INF).-e [STRING]: Path of the mutations' name file. If this parameter is not used then the mutation will be named progressively from1.-E [STRING]: Path of the cells' name file. If this parameter is not used then the cells will be named progressively from1.-g [FLOAT/STRING]: Loss rate in the input file or path of the file containing different GAMMA rates for each mutations.-q [FLOAT/STRING]: Recurrent rate in the input file or path of the file containing different DELTA rates for each mutations.-R [INT]: Set the total number of Simulated Annealing repetitions. Default is 5.-M: Force reSASC to infer a monoclonal tree, i.e. a tree with only one node child of the germline. Default is not set.
Output parameters (optional)
-l: If this flag is used SASC will output a mutational tree with cells attached to it. Otherwise cells will not present. Please note that this flag is needed if you plan to run the visualization tool afterwards.-x: If this flag is used, SASC will additionally output the expected matrix E.-P [STRING]: Prefix for the output files.
Simulated Annealing parameters (optional)
-S [FLOAT]: Starting temperature of the Simulated Annealing algorithm.-C [FLOAT]: Cooling rate of the Simulated Annealing algorithm.-p [INT]: Total number of cores to be used by the tool.
Error learning parameters (optional)
-A [FLOAT]: Standard deviation for new FN discovery. [Disabled by default.]-B [FLOAT]: Standard deviation for new FP discovery. [Disabled by default.]-G [FLOAT]: Standard deviation for new GAMMA discovery. [Disabled by default.]-Q [FLOAT]: Standard deviation for new DELTA discovery. [Disabled by default.]
reSASC has three different output formats that can be toggled with different arguments.
Mutational Tree
This is the standard output; reSASC will generate a mutational tree in DOT format with no cells attached as leaves of the tree. An example of this output is shown in MGH36 mutational tree.
Mutational Tree with cells as leaves
By toggling option -l reSASC will instead output a mutational tree in DOT format with cells attached as leaves of the tree. An example can be found at MGH36 mutational tree with cells.
Expected Matrix
In addition to the previous formats SASC can output the expected matrix E such in MGH36 expected matrix using the -x flag.
SASC can then be run using the previously described parameters. Here we show a list of run and their results.
Childhood Lymphoblastic Leukemia (patient 4)
./sasc -i data/real/gawad/pat4.txt -m 78 -n 143 -a 0.3 -b 0.001 -k 3 -z 5 -e data/real/gawad/pat4_mut.txt The command specifies a Dollo-3 phylogeny with a maximum of 5 deletions in the tree, a single FN rate of 0.3, no prior loss probability (default to 1) and mutations names specified in the file data/real/gawad/pat4_mut.txt.
MGH36 with different FN rates and monoclonality
./sasc -i data/real/MGH36/MGH36_scs.txt -m 77 -n 579 -a data/real/MGH36/MGH36_fn-rates.txt -b 0.005 -k 0 -e data/real/MGH36/MGH36_snv-names.txt -E data/real/MGH36/MGH36_cell-names.txt -l -x -R 1 -MThe command specifies a Perfect Phylogeny (Dollo-0) with FN rates detailed in file data/real/MGH36/MGH36_fn-rates.txt, mutation names in data/real/MGH36/MGH36_snv-names.txt, cell names in data/real/MGH36/MGH36_cell-names.txt, output of mutational tree with cells as leaves (-l), output of the expected matrix (-x) and a total of 1 repetition (-r 1).
Simulation with different FN rates and Prior values and Error Learning
./sasc -i data/simulated/exp6-bimod/sim_21_scs.txt -m 50 -n 200 -k 1 -d 3 -a examples/alphas.txt -g examples/gammas.txt -b 0.0003 -A 0.2 -G 0.05Simulation with different FN rates and Prior values and Error Learning and recurrent mutations
./sasc -i data/simulated/exp6-bimod/sim_21_scs.txt -m 50 -n 200 -k 1 -r 2 -R 2 -d 3 -c 4 -a examples/alphas.txt -g examples/gammas.txt -d examples/deltas.txt -b 0.0003 -A 0.2 -G 0.05 -D 0.02The command specifies a Dollo-1 phylogeny with a maximum of 3 deletions in the tree, FN rates detailed in examples/alphas.txt, prior loss probabilities in examples/gammas.txt, learning standard deviation of 0.2 for FN rate (-A 0.2) and learning standard deviation of 0.05 for prior loss (-G 0.05).
The script SASC-viz.py can be used as a visualization tool and to apply operation to reSASC's output without needing to re run the tool. The tool will change the output of the tool only for visualization purposes. Please note that you have to run reSASC with the -l flag in order to use the tool.
python3 SASC-viz.py [-h] -t TREE [-E CELLNAMES | -n TOTCELL] [--show-support]
[--show-color] [--collapse-support COLLAPSE_SUPPORT]
[--collapse-simple] [--sep SEP]Input files required
-t TREE: path to the input file, i.e.SASC's output (run with the-lflag).
You are required to use either:
-E CELLNAMES: path to the mutation labels file (this is the same file used with the flag-EonSASC).-n TOTCELLS: instead of the previous argument, if the mutation labels are not specified, you must provide the number of cells in the input (this is the same option used with-nwithSASC).
Supported operations (optional)
--collapse-simple: when this option is activated, all simple non-branching paths are collapsed, i.e. if a node has only one child, then such node is merged with its child.--show-support: show the mutation support of each node.--collapse-support THRESHOLD: if the support of a node x is lower than the specifiedTHRESHOLD, then x is merged with its parent. This operation is performed in a bottom-up fashion starting from the leaves.--show-color: if this flag is used, the nodes will be colored using a gradient scale from red to green based on the support.--sep STRING: if this option is used the labels on the nodes will be separated bySTRING. Default is,.
Output
The tool will output a tree in DOT format, which can be converted into a picture using either the program dot or the web interface available on webgraphviz.com.
Run SASC-viz on the MGH36 dataset, run with the parameters used in the previous example.
python3 SASC-viz.py -t examples/MGH36_scs_mlt.gv -E data/real/MGH36/MGH36_cell-names.txt --collapse-simple --collapse-support 20 --show-supportOutput:
digraph phylogeny {
node [penwidth=2];
"0" [label="germline [255 cells]"];
"0" -> "1";
"1" [label="IDH1,NOTCH2,RTTN,TBC1D10A,MLYCD,CACNA1G,CTNNA2,NRN1,APC2,IL33,NBPF10,RFX3,UBE2Z,ZZEF1,KHSRP,SH3BP5,CCDC181,VGLL4,PIK3CA,PHLDB3,NR3C1,RP11-356C4.3,VPS9D1,PLEKHM1,LINC00937,ST8SIA3,CPEB4,TRPM3,TRIOBP,ZNF451,CEP55,TFAP2A,ZNF721,KIF2A,USP36,IFT81,SVEP1,MCM8,ARHGEF3,AGAP2,NR5A2 [s=100%]"];
"1" -> "23";
"23" [label="CEBPZ,DGCR6L,MAN1B1,ENO3,ZNF526 [s=76%]"];
"23" -> "25";
"25" [label="MIR4477B,KMT2C,SLC26A11,ORC3,CLEC18B,KAT6A,CNNM2,SLC16A7 [s=100%]"];
"25" -> "27";
"27" [label="PCDHA1 [s=28%]"];
"27" -> "28";
"28" [label="HELZ2,RIN2 [s=50%]"];
"27" -> "30";
"30" [label="TXNDC2,HEATR4 [s=25%]"];
"27" -> "32";
"32" [label="NPEPL1 [s=25%]"];
"25" -> "33";
"33" [label="EEF1B2,ZNF462,EP400,RP11-403I13.8 [s=71%]"];
"1" -> "46";
"46" [label="HLA-DQB2,ABCA7,STXBP1,RUNX2,SOX5,KIAA0907,CPAMD8 [s=23%]"];
"46" -> "47";
"47" [label="ANKRD30B,FAM182B,TRPM2,AS3MT [s=25%]"];
"46" -> "52";
"52" [label="EMR2,CYP27A1 [s=75%]"];
}
Run SASC-viz to produce directly a PDF file containing the tree. This requires dot to be installed.
python3 SASC-viz.py -t examples/simulated_mlt.gv -n 50 --collapse-simple --collapse-support 5 --show-support | dot -Tpdf > tree.pdfOutput:
The support si of a mutation i is computed on the n x m inferred matrix E as follows. Let pr(i) be the set of nodes in the path from the root to i let st(i) be the set of nodes in the subtree rooted in i, and let C(i) be the number of cells assigned to the node i. Then the mutation support si is:
The paper's experiments can be reproduced following instruction in the supplementary repository