🚀🚀🚀 A TensorFlow implementation of BiLSTM+CRF model, for sequence labeling tasks.
- based on Tensorflow api.
- highly scalable; everything is configurable.
- modularized with clear structure.
- very friendly for beginners.
- easy to DIY.
Sequential labeling is one typical methodology modeling the sequence prediction tasks in NLP.
Common sequential labeling tasks include, e.g.,
- Part-of-Speech (POS) Tagging,
- Chunking,
- Named Entity Recognition (NER),
- Punctuation Restoration,
- Sentence Boundary Detection,
- Scope Detection,
- Chinese Word Segmentation (CWG),
- Semantic Role Labeling (SRL),
- Spoken Language Understanding,
- Event Extraction,
- and so forth...
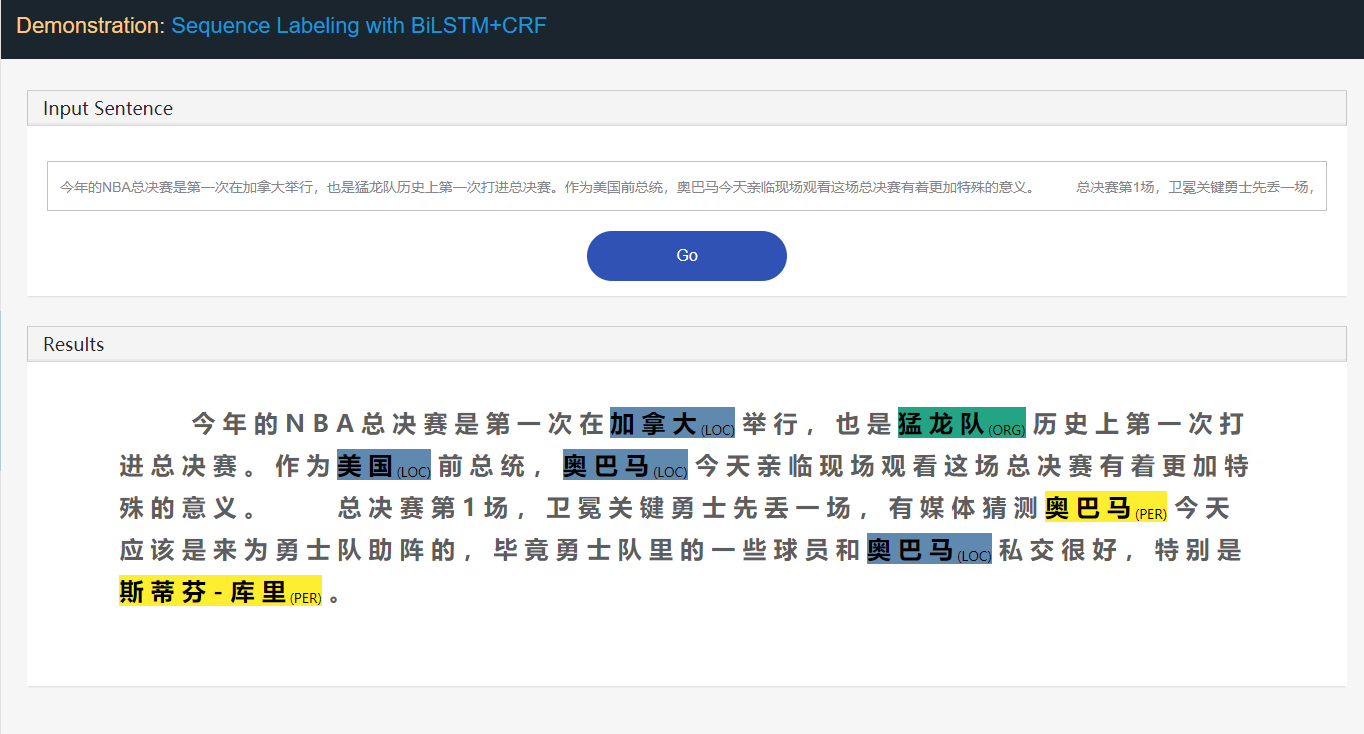
Taking Named Entity Recognition (NER) task as example:
Stanford University located at California .
B-ORG I-ORG O O B-LOC Ohere, two entities, Stanford University and California are to be extracted.
And specifically, each token in the text is tagged with a corresponding label.
E.g., {token:Stanford, label:B-ORG}.
The sequence labeling model aims to predict the label sequence, given a token sequence.
BiLSTM+CRF proposed by Lample et al., 2016, is so far the most classical and stable neural model for sequential labeling tasks.
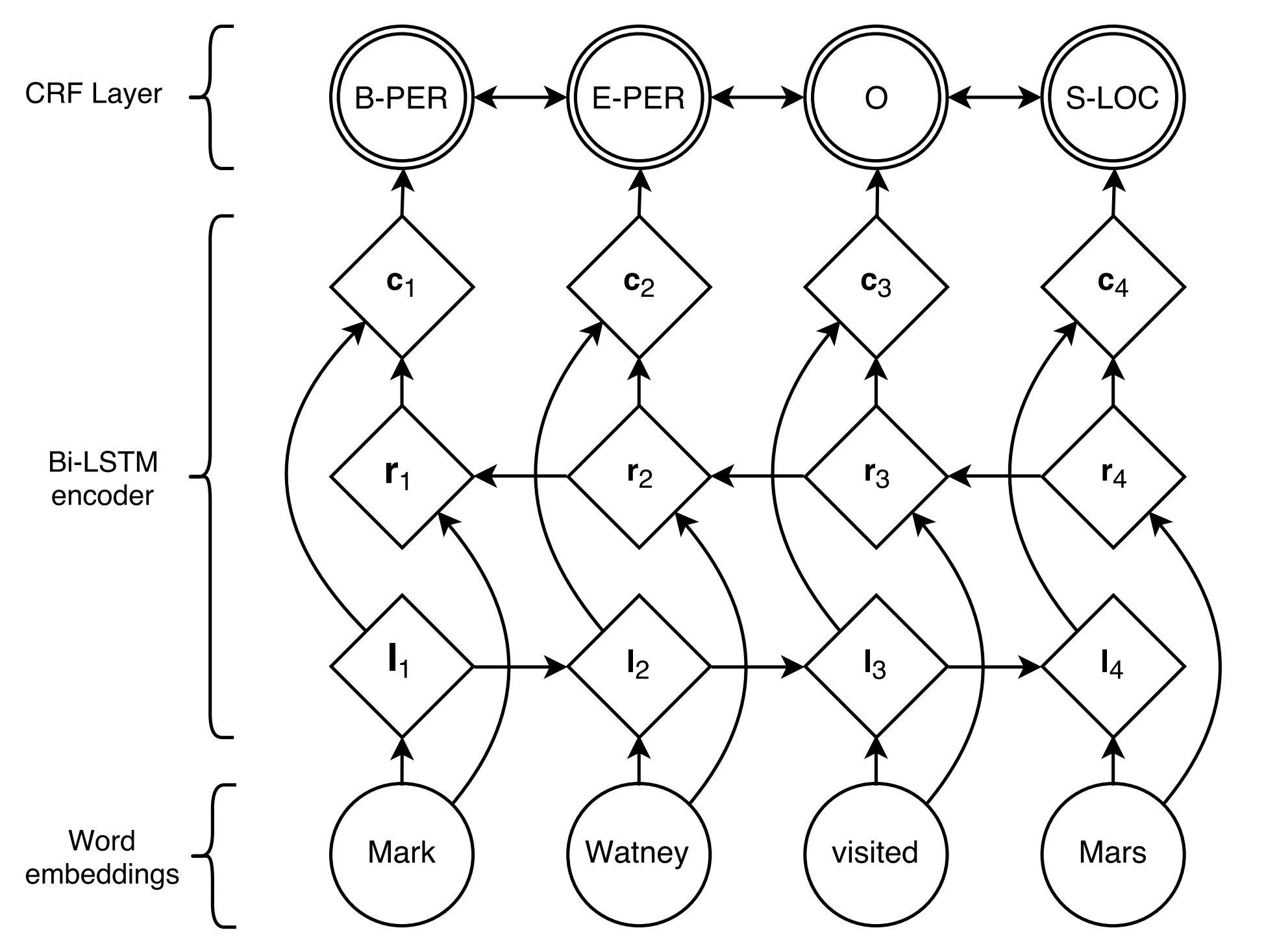
-
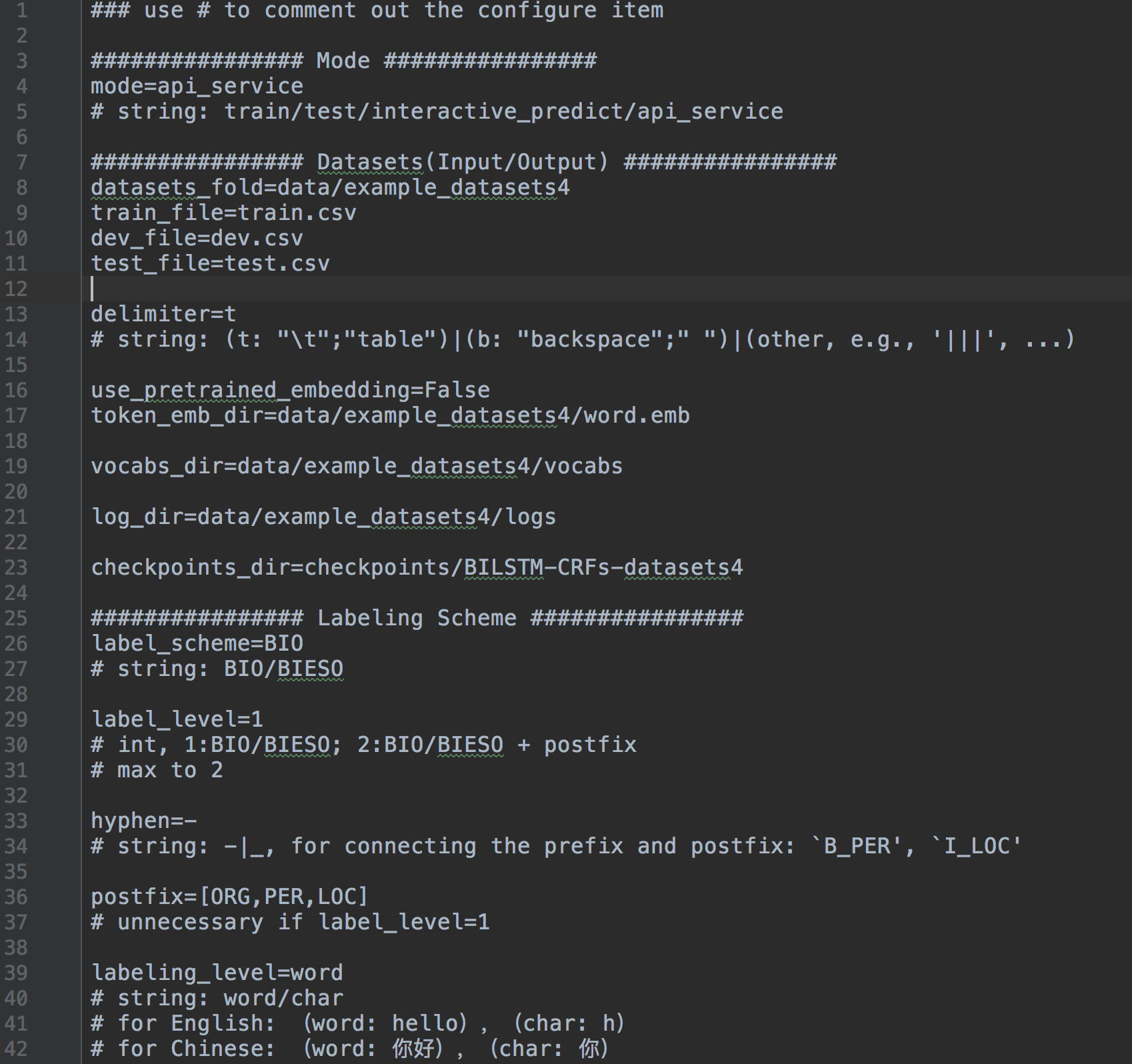
configuring all settings
- Running Mode: [
train/test/interactive_predict/api_service] - Datasets(Input/Output):
- Labeling Scheme:
- [
BIO/BIESO] - [
PER|LOC|ORG] - ...
- [
- Model Configuration:
- encoder: BGU/Bi-LSTM, layer, Bi/Uni-directional
- decoder: crf/softmax,
- embedding level: char/word,
- with/without self attention
- hyperparameters,
- ...
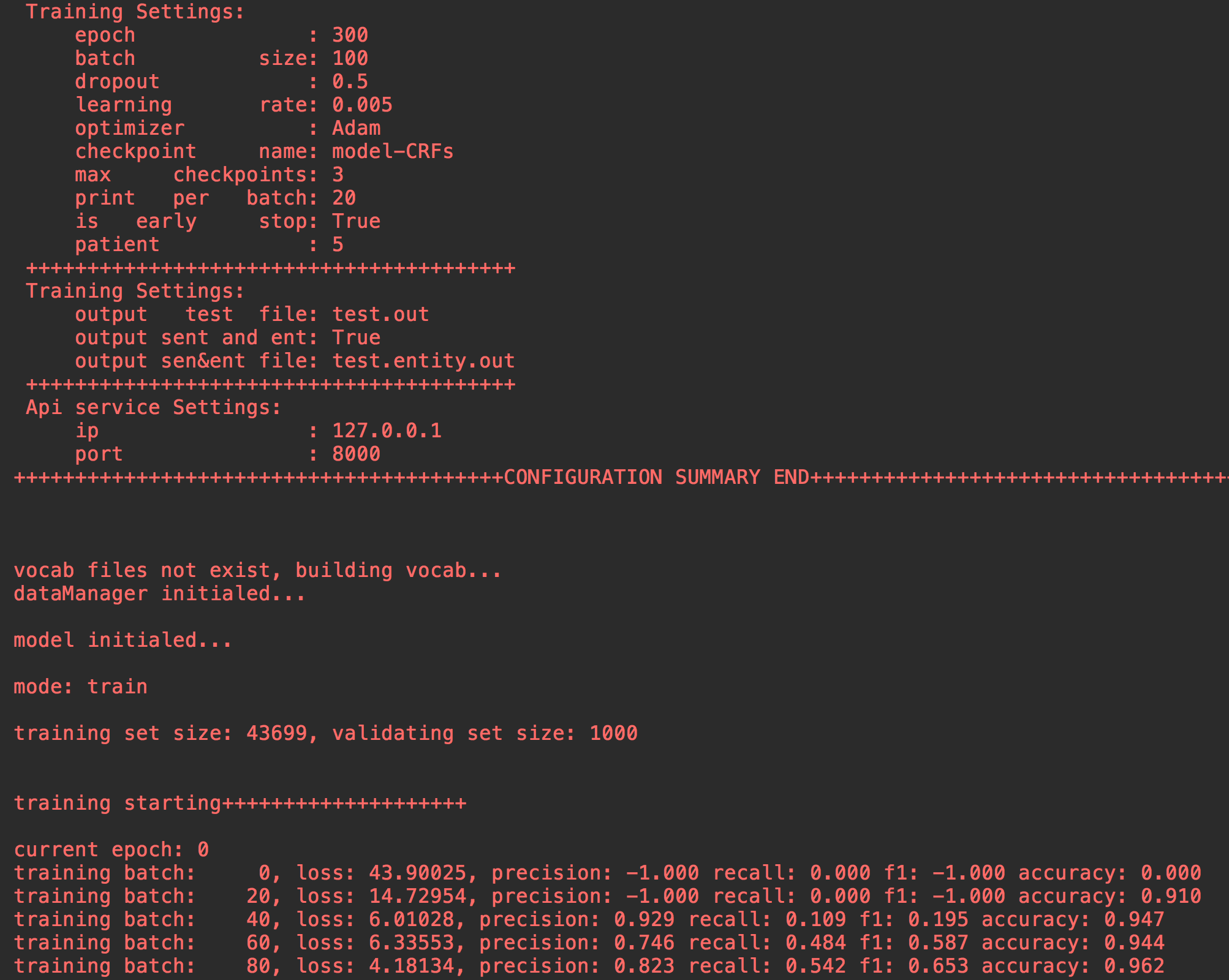
- Training Settings:
- subscribe measuring metrics: [precision,recall,f1,accuracy]
- optimazers: GD/Adagrad/AdaDelta/RMSprop/Adam
- Testing Settings,
- Api service Settings,
- Running Mode: [
-
logging everything
-
web app demo for easy demonstration
-
object oriented: BILSTM_CRF, Datasets, Configer, utils
-
modularized with clear structure, easy for DIY.
see more in HandBook.
- python >=3.5
- tensorflow >=1.8
- numpy
- pandas
- Django==1.11.8
- jieba
- ...
download the repo for directly use.
git clone https://github.com/scofield7419/sequence-labeling-BiLSTM-CRF.git
pip install -r requirements.txt
install the BiLSTM-CRF package as a module.
pip install BiLSTM-CRF
usage:
from BiLSTM-CRF.engines.BiLSTM_CRFs import BiLSTM_CRFs as BC
from BiLSTM-CRF.engines.DataManager import DataManager
from BiLSTM-CRF.engines.Configer import Configer
from BiLSTM-CRF.engines.utils import get_logger
...
config_file = r'/home/projects/system.config'
configs = Configer(config_file)
logger = get_logger(configs.log_dir)
configs.show_data_summary(logger) # optional
dataManager = DataManager(configs, logger)
model = BC(configs, logger, dataManager)
###### mode == 'train':
model.train()
###### mode == 'test':
model.test()
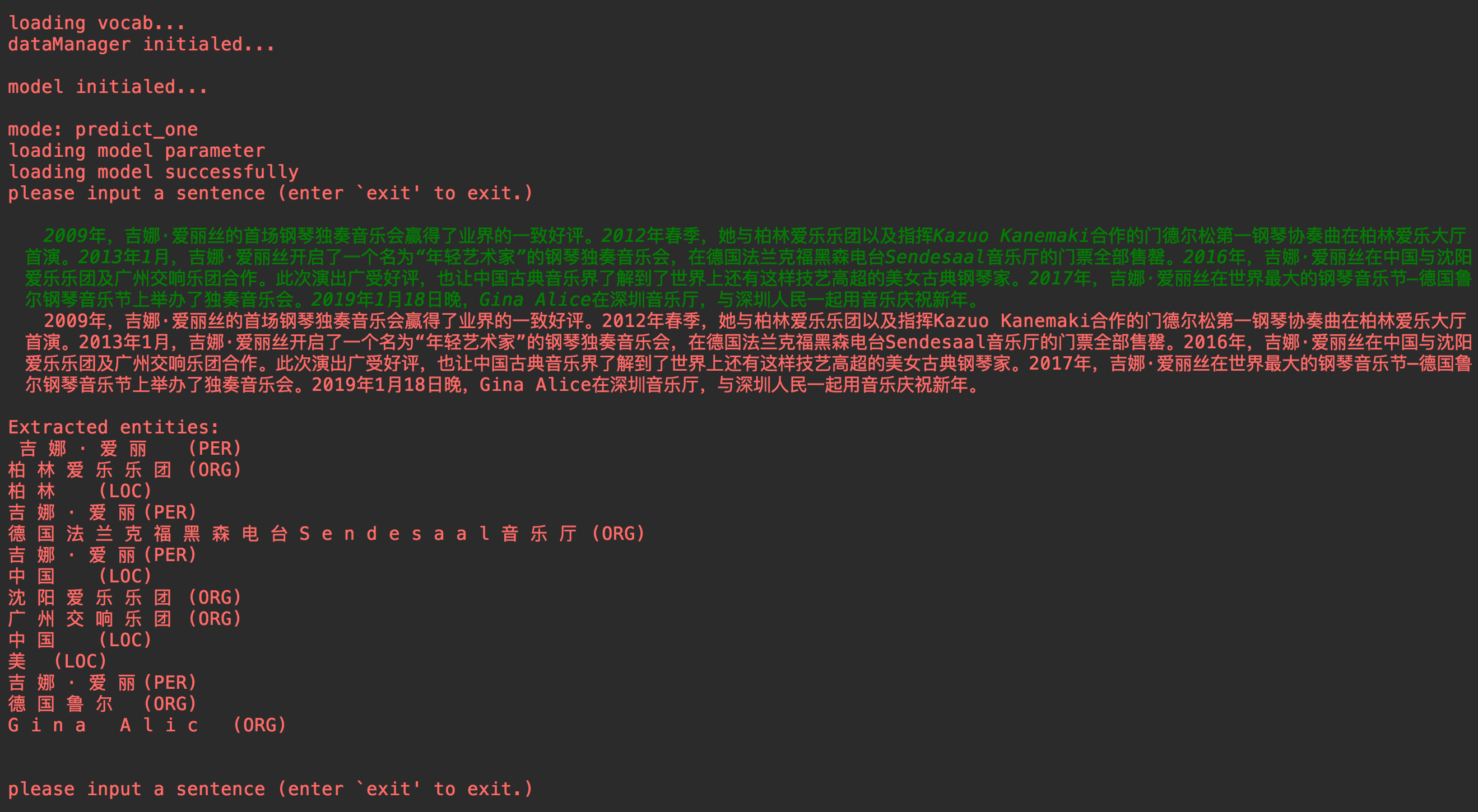
###### mode == 'single predicting':
sentence_tokens, entities, entities_type, entities_index = model.predict_single(sentence)
if configs.label_level == 1:
print("\nExtracted entities:\n %s\n\n" % ("\n".join(entities)))
elif configs.label_level == 2:
print("\nExtracted entities:\n %s\n\n" % ("\n".join([a + "\t(%s)" % b for a, b in zip(entities, entities_type)])))
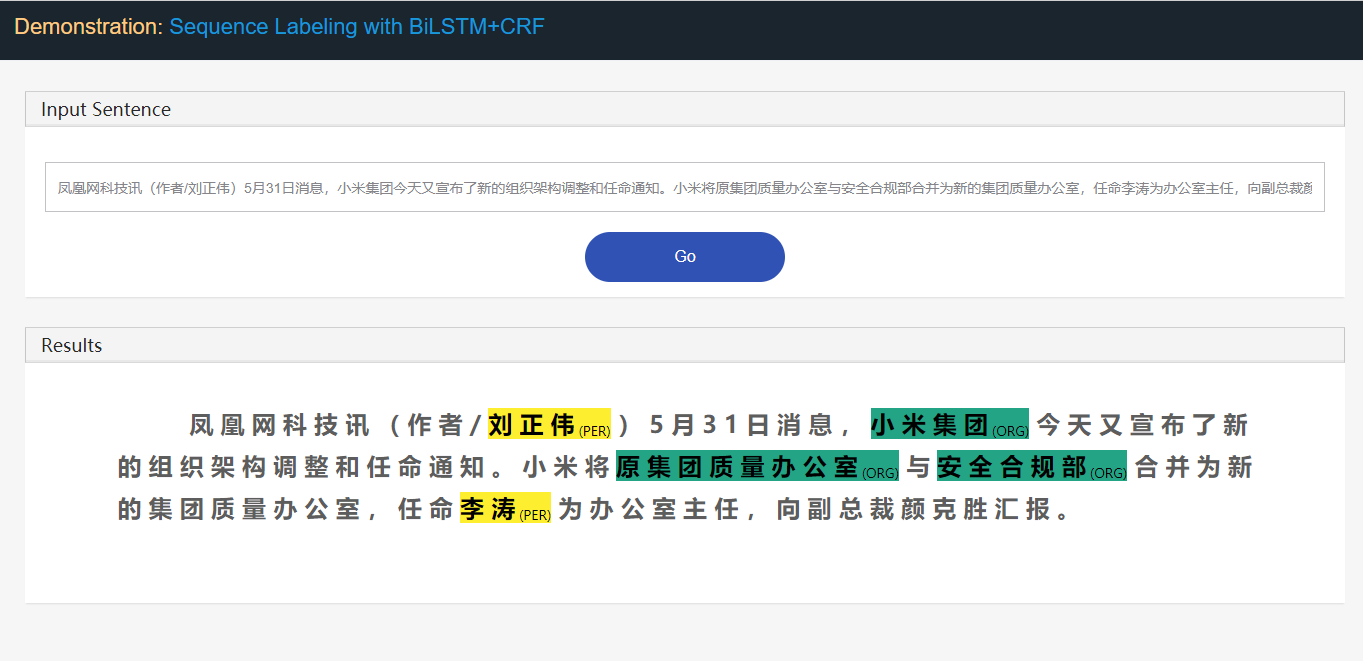
###### mode == 'api service webapp':
cmd_new = r'cd demo_webapp; python manage.py runserver %s:%s' % (configs.ip, configs.port)
res = os.system(cmd_new)
open `ip:port` in your browser.
├── main.py
├── system.config
├── HandBook.md
├── README.md
│
├── checkpoints
│ ├── BILSTM-CRFs-datasets1
│ │ ├── checkpoint
│ │ └── ...
│ └── ...
├── data
│ ├── example_datasets1
│ │ ├── logs
│ │ ├── vocabs
│ │ ├── test.csv
│ │ ├── train.csv
│ │ └── dev.csv
│ └── ...
├── demo_webapp
│ ├── demo_webapp
│ ├── interface
│ └── manage.py
├── engines
│ ├── BiLSTM_CRFs.py
│ ├── Configer.py
│ ├── DataManager.py
│ └── utils.py
└── tools
├── calcu_measure_testout.py
└── statis.py
-
Folds
- in
enginesfold, providing the core functioning py. - in
data-subfoldfold, the datasets are placed. - in
checkpoints-subfoldfold, model checkpoints are stored. - in
demo_webappfold, we can demonstrate the system in web, and provides api. - in
toolsfold, providing some offline utils.
- in
-
Files
main.pyis the entry python file for the system.system.configis the configure file for all the system settings.HandBook.mdprovides some usage instructions.BiLSTM_CRFs.pyis the main model.Configer.pyparses thesystem.config.DataManager.pymanages the datasets and scheduling.utils.pyprovides on the fly tools.
Under following steps:
- configure the Datasets(Input/Output).
- configure the Labeling Scheme.
- configure the Model architecture.
- configure the webapp setting when demonstrating demo.
- configure the running mode.
- configure the training setting.
- run
main.py.
- configure the running mode.
- configure the testing setting.
- run
main.py.
- configure the running mode.
- run
main.py. - interactively input sentences.
- configure the running mode.
- configure the api_service setting.
- run
main.py. - make interactively prediction in browser.
Datasets including trainset, testset, devset are necessary for the overall usage. However, is you only wanna train the model the use it offline, only the trainset is needed. After training, you can make inference with the saved model checkpoint files. If you wanna make test, you should
For trainset, testset, devset, the common format is as follows:
- word level:
(Token) (Label)
for O
the O
lattice B_TAS
QCD I_TAS
computation I_TAS
of I_TAS
nucleon–nucleon I_TAS
low-energy I_TAS
interactions E_TAS
. O
It O
consists O
in O
simulating B_PRO
...
- char level:
(Token) (Label)
马 B-LOC
来 I-LOC
西 I-LOC
亚 I-LOC
副 O
总 O
理 O
。 O
他 O
兼 O
任 O
财 B-ORG
政 I-ORG
部 I-ORG
长 O
...
Note that:
- the
testsetcan only exists with the theTokenrow. - each sentence of tokens is segmented with a blank line.
- go to the example dataset for detailed formation.
During testing, model will output the predicted entities based on the test.csv.
The output files include two: test.out, test.entity.out(optional).
-
test.outwith the same formation as input
test.csv. -
test.entity.out
Sentence
entity1 (Type)
entity2 (Type)
entity3 (Type)
...
If you wanna adapt this project to your own specific sequence labeling task, you may need the following tips.
-
Download the repo sources.
-
Labeling Scheme (most important)
- label_scheme: BIO/BIESO
- label_level: with/without suffix
- hyphen, for connecting the prefix and suffix:
B_PER',I_LOC' - suffix=[NR,NS,NT]
- labeling_level: word/char
-
Model: modify the model architecture into the one you wanted, in
BiLSTM_CRFs.py. -
Dataset: adapt to your dataset, in the correct formation.
-
Training
- specify all directories.
- training hyperparameters.
For more useage details, please refers to the HandBook
You're welcomed to issue anything wrong.
- 2019-Jun-04, Vex version, v1.0, supporting configuration, scalable.
- 2018-Nov-05, support char and word level embedding.
- 2017-Dec-06, init version, v0.1.