A database of global locales to support modeling and simulation in epidemiology with the current focus on the COVID 19 pandemic.
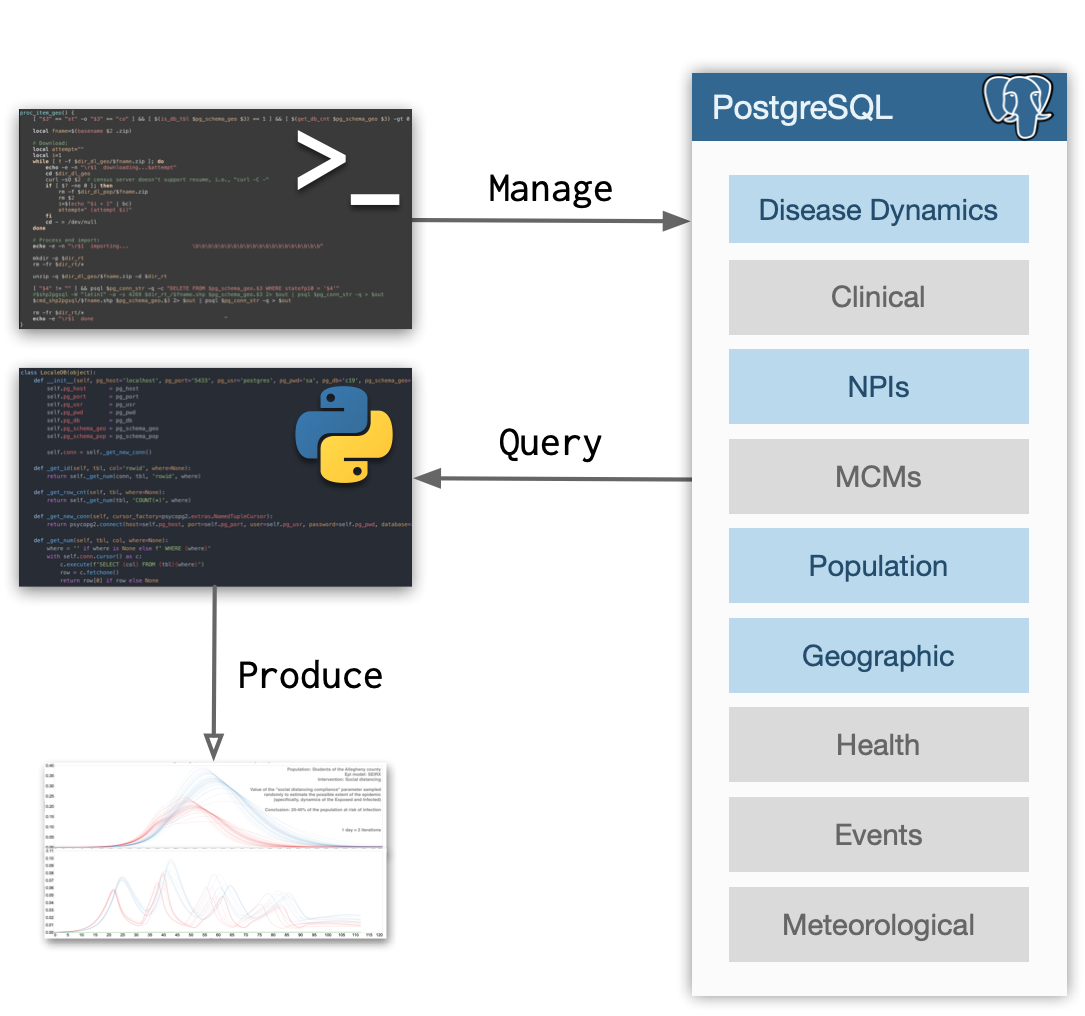
As shown on the figure below, LocaleDB stores several types of data (gray boxes indicate planned future extentions). That data is stored in a PostgreSQL database which is managed by a command line tool (localedb) and a Python script (localedb_man.py). The content of the database is accessed via a Python package which provides a high level API to, for example, suggest U.S. counties similar to the county specified.
This design that separates data management and data consumption reflects the anticipated production use case. Namely, the database will be deployed and set up once and will then require little to no manual management (periodic updates will be autonomous). It will then be used for producing data that will drive modeling and simulation efforts.
As depicted on the figure above, the current projection is for LocaleDB to contain the following data types:
- Disease dynamics (e.g., number of confirmed cases)
- Clinical (e.g., R0, incubation period, proportion of asymptomatic cases, etc.)
- Non-pharmaceutical interventions (NPIs; e.g., dates of stay-at-home order)
- Medical countermeasures (MCMs; e.g., vaccine availability, efficacy, and allocation strategies)
- Population (e.g., households, their incomes, age of people, etc.)
- Geographic and cartographic (e.g., area of land, population density) -Mobility (mobile-phone based)
- Health factors and outcomes (e.g., diet, exercise, access to care, etc.)
- Local events (e.g., dates and sizes of mass protests)
- Meteorological
All that data will be stratified by locale at all available levels of spatial aggregation (e.g., country, state, county, tract, block group, block). In terms of temporal resolution, the highest frequency with which processes are sampled/measured will be the goal. For example, disease dynamics will be represented as a time series of daily numbers of confirmed cases and deaths, while health factors and outcomes will be encoded with far fewer time steps (probably months).
LocaleDB can be deployed to a development and production environments. It is recommended to familiarize yourself with the software using the development environment first.
- curl or wget
- Docker
- curl or wget
- PostgreSQL client
- PostgreSQL server (with PostGIS and TimescaleDB extensions)
- Python 3
Note: LocaleDB should not be deployed to a production environment yet. This note will be removed when that deployment mode has been fully implemented and fully tested.
On MacOS run:
sh -c "$(curl -fsSL https://raw.githubusercontent.com/momacs/localedb/master/setup.sh -O -)"
On Linux run:
sh -c "$(wget -q https://raw.githubusercontent.com/momacs/localedb/master/setup.sh -O -)"
Alternatively, you can run the commands from the setup.sh script manually.
Production environment: For production deployment, after the installation script above has finished, edit the $HOME/bin/localedb script and change is_prod=0 to is_prod=1. This step is left to be done manually to ensure intent.
It is never a bad idea to first create a new Python virtual environment:
# sudo apt install python3-venv # may be needed on Linux
python3 -m venv ./prj01
cd prj01
source ./bin/activate
Then, install the package like so:
pip install git+https://github.com/momacs/localedb.git
After setting up the command line management tool, setup the LocaleDB instance:
$ localedb setup
Initializing data structures... done
Loading locales... done
To display filesystem information, run:
$ localedb info fs
Directory structure
Root /Users/tomek/.localedb 43M
Runtime /Users/tomek/.localedb/rt 0B
PostgreSQL data /Users/tomek/.localedb/pg 43M
Disease data /Users/tomek/.localedb/dl/dis 0B
Geographic data /Users/tomek/.localedb/dl/geo 0B
Population data /Users/tomek/.localedb/dl/pop 0B
To import COVID-19 disease data (currently only dynamics and non-pharmaceutical interventions), run:
$ localedb import dis c19
Disease dynamics
Loading global confirmed... done (11 s)
Loading global deaths... done (14 s)
Loading global recovered... done (12 s)
Loading US confirmed... done (141 s)
Loading US deaths... done (155 s)
Consolidating... done (88 s)
Non-pharmaceutical interventions
Loading Keystone... done (14 s)
To see some basic database statistics, run:
$ localedb info data
Data
Main
Locale count 4153
Country count 188
Disease (c19)
Dynamics
Locale count 3607
Date range 2020-01-22 2020-09-17
Observation count 865680
Observation count per locale 240.00 (SD=0.00)
Non-pharmaceutical interventions
Locale count 669
Data range 2010-04-27 2020-07-27
NPI count 5162
NPI count per locale 7.72 (SD=1.57)
Count per type
669 school closure
667 closing of public venues
666 non-essential services closure
637 shelter in place
622 gathering size 10 0
582 social distancing
471 religious gatherings banned
406 gathering size 100 26
278 gathering size 500 101
132 gathering size 25 11
32 lockdown
...
To import geographic and cartographic data for the state of Alaska, run:
$ localedb load geo AK
US states done
US counties done
AK tracts done
AK block groups done
AK blocks done
Analyzing database... done
To import synthetic population data, run:
$ localedb load pop AK
AK done
Analyzing database... done
Imported states can be removed like so:
localedb db rm state-geo AK
localedb db rm state-pop AK
localedb db rm state AK # remove all data types
Once data has been imported and the downloaded data files are no longer needed, they can be removed like so:
localedb fs rm-data geo
localedb db rm-data pop
localedb db rm data-all # remove all data files
To stop LocaleDB instance, run:
localedb stop
To uninstall LocaleDB (leaving nothing behind), run:
localedb uninstall
For the list of available commands, run localedb. For an explanation of each command, run localedb help. Keep in mind that some commands have subcommands.
Here is an example of the Python LocaleDB package can be used:
from localedb import LocaleDB
db = LocaleDB()
db.set_pop_view_household('02') # constrain view to households located in Alaska
print(db.get_pop_size()) # get size of population that lives in those households
db.set_pop_view_household('02013') # do the same for one of the counties in Alaska
print(db.get_pop_size())If the database is not installed on the localhost (or if any other connection parameters need to be adjusted), they should be passed to the LocaleDB class' constructor. Documentation of the package will be published later on.
- COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University
- Keystone: COVID-19 Intervention Data
- 2010 U.S. Synthesized Population Dataset
- US Census Bureau: TIGER/Line Shapefiles (2010)
This project is licensed under the BSD License.