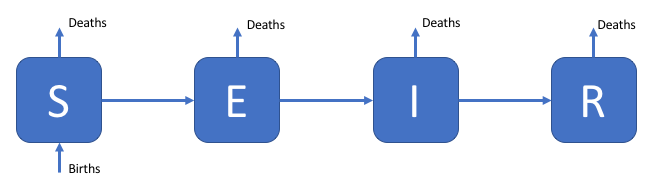
This shiny app has been developed to allow the exploration of the parameter space of compartmental infectious disease models. It is designed to be used as a teaching aid when introducing people to the concepts behind infectious disease models without requiring them to interact with the underlying code. A number of infectious disease models are included, such as a Susceptible, Exposed, Infected, Recovered model the flow diagram for which can be seen below.
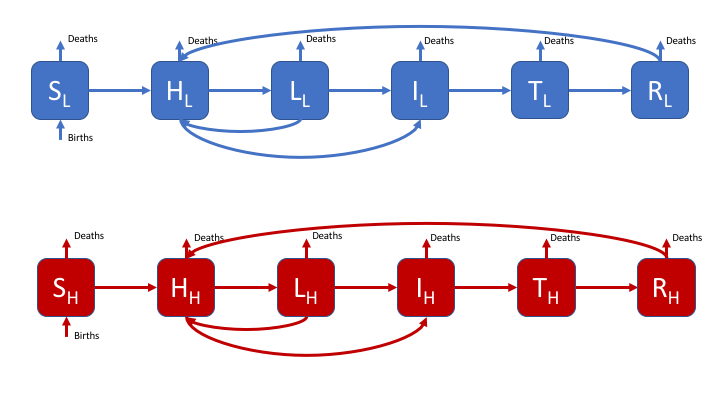
An example of a more complex model for Tuberculosis is also included,
This includes a risk group, high and low risk latent states, reinfection
for those who have recovered from active disease or are latently
infected, and treatment. The impact of demographic processes are
explored in all models. The code for each model can be viewed in the
application. All functionality seen in this app is provided by
idmodelr.
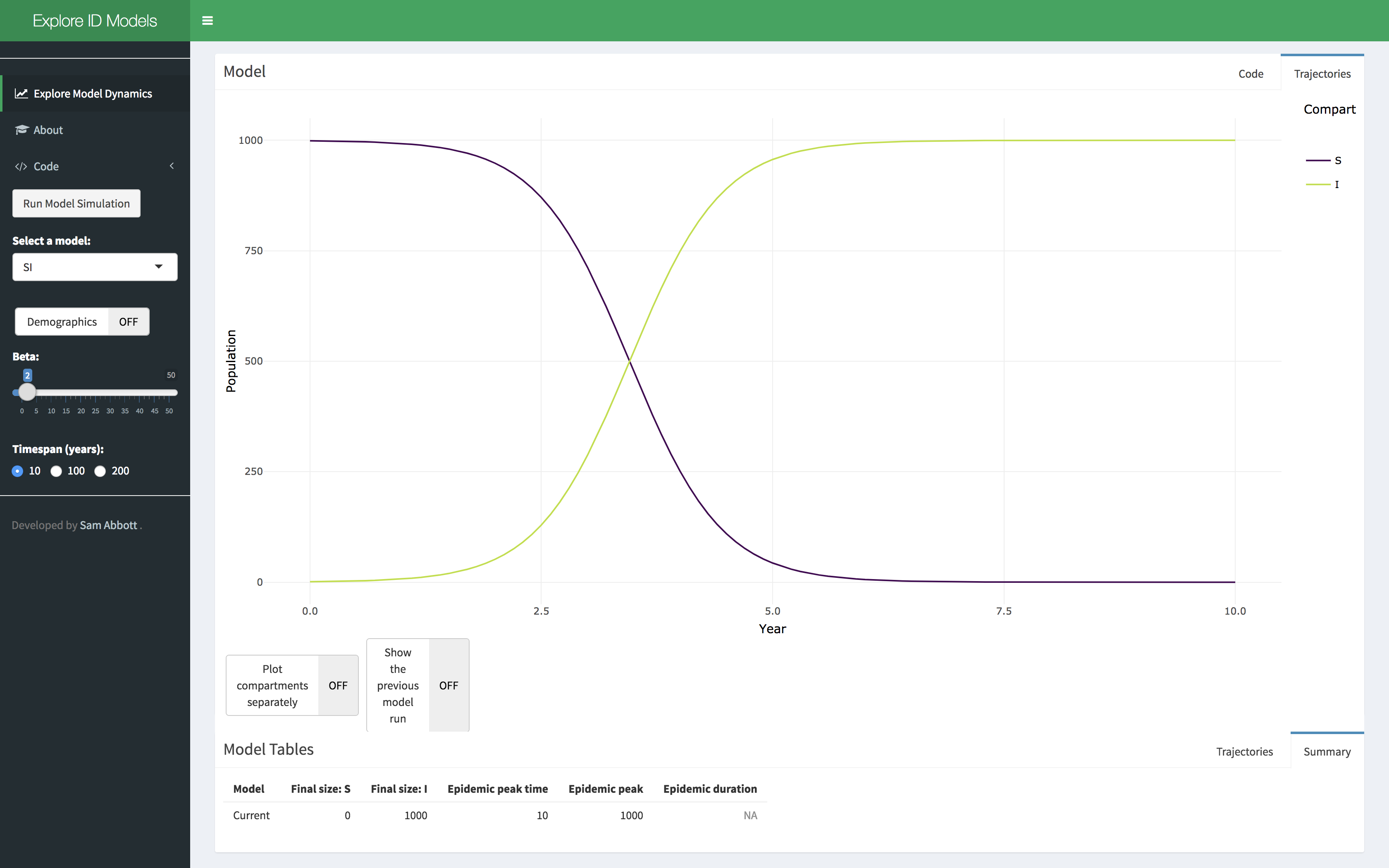
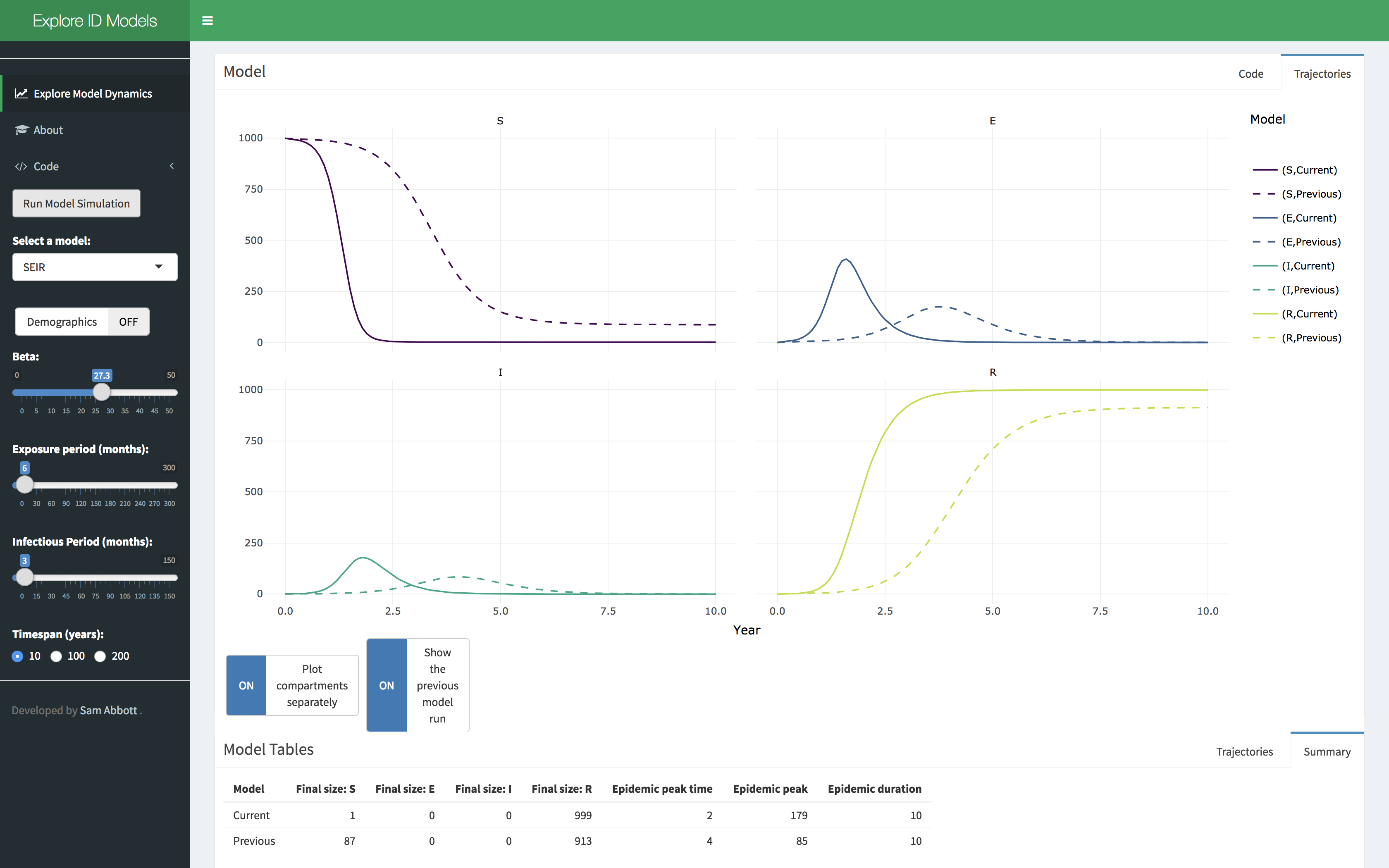
- Click on the Explore Model Dynamics Tab in the menu on the left hand side. You should see this,
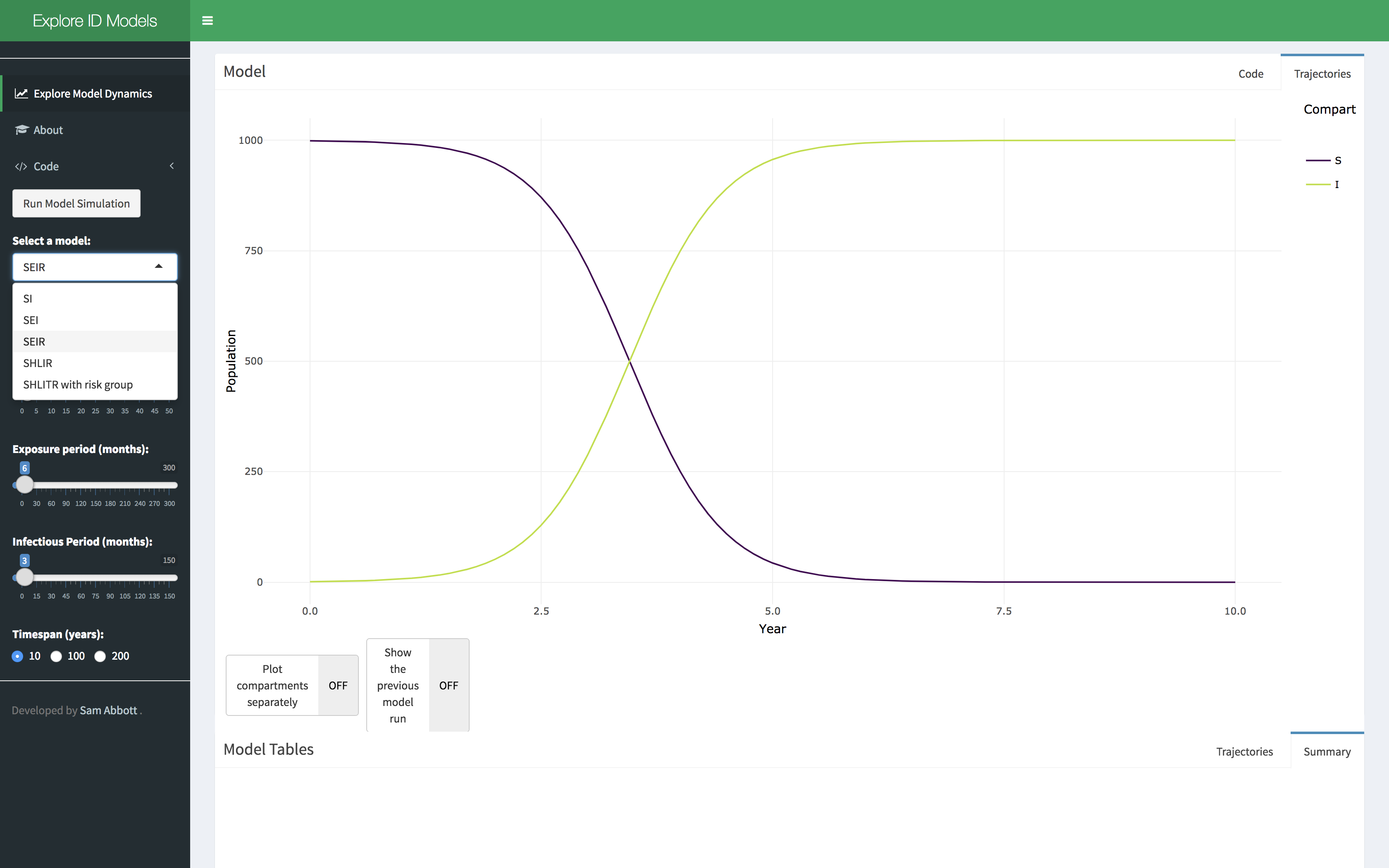
- Select a model of interest, demographic processes can be toggled on or off. Click on the run model simulation button to simulate the model.
- Explore the parameter space of the model using the provided sliders. A summary table of the model simulation is provided as are the raw simulation results. Show the previous model simulation using the previous model run slider. The model output can also be shown separately for each compartment using the plot compartments separately slider.
The app can be run directly from GitHub. Open R and run the following;
install.packages("shiny")
shiny::runGitHub("exploreidmodels", "seabbs")If the above fails the likely reason is that a required package has failed to install. See here for a list of required packages.
To install and run the shiny app locally on your own computer you will
need to first install R, it is also
suggested that you install
Rstudio. After
downloading the source code from this
repository click on the
exploreidmodels.Rprof file, this will open an Rstudio window. Type the
following code into the command line;
install.packages("packrat")
packrat::restore()To run the app open the ui.R file and press run, depending on your
computer this may take some time.
Docker is a container software that seeks to eliminate “works on my machine” issues. For installation and set up instructions see here.
This docker container is based on the tidyverse docker image, see here for instructions on use. To run the docker image run the following in a bash shell:
docker pull seabbs/exploreidmodels
docker run --rm -p 8787:8787 -e USER=exploreidmdoels -e PASSWORD=exploreidmodels --name exploreidmodels seabbs/exploreidmodelsThe Rstudio server can be found on port :8787 at your local machines
ip (or localhost on windows). Alternatively the app has been deployed
using mybinder. See
here
for the development environment and
here
for the app.