This repository has several codes to calculate the potential of mean force (pmf) based on radial distribution functions (rdf) of protein-protein interaction. The input are rdf files generated with a modified version of Faunus packages (https://github.com/mlund/faunus). Codes were employed to calculate pmf curves in the publication: https://doi.org/10.1016/j.virusres.2022.198838. Here is showing an example simplified!
Unzip the file 'DATA.tar.gz'.
tar -xzvf DATA.tar.gz
Notes:
- We strictly recommend using the same composition of folders and subfolders as the 'DATA' directory.
Start cleaning the directory.
chmod +x z.clean.sh ; ./z.clean.sh ; chmod -x z.clean.sh
Evaluation of aam used for simulation to detect match with the correct aam structure.
echo 1.aam_data.sh
chmod +x ./1.aam_data.sh
./1.aam_data.sh > 1.aam_data.log
- The script will generate an output called 1.aam_data.log, where it will be shown if the structure is different ("YES") or not ("NO").
Combine the rdf files in a unique directory. During the calculation just curves greater than specific number of point will be considered.
Notes:
- The "minimum distance" in 2.combine_data.log should be an int number.
- The script should assign to all rdf files the same number of row, else something is wrong! Pay attetion to it!!! All next calculation depends on that.
- The script depends on the previous output 1.valid_data.txt.
echo 2.combine_data.sh
chmod +x ./2.combine_data.sh
./2.combine_data.sh > 2.combine_data.log
- The output generated 2.total_number_curves.log will show the total number of curves per condition.
- The output 2.curves_bad.dat will show curves with problems of sampling.
Convert rdf to pmf (3.pmf.py), compare curves (3.comparison.py), and remove redudant information (3.remove_rep.sh).
echo 3.convert_rdf2pmf.sh
chmod +x 3.convert_rdf2pmf.sh
./3.convert_rdf2pmf.sh > 3.convert_rdf2pmf.log # Will call the three different scripts.
echo 4.calculate_average.sh
chmod +x 4.calculate_average.sh
./4.calculate_average.sh > 4.calculate_average.log
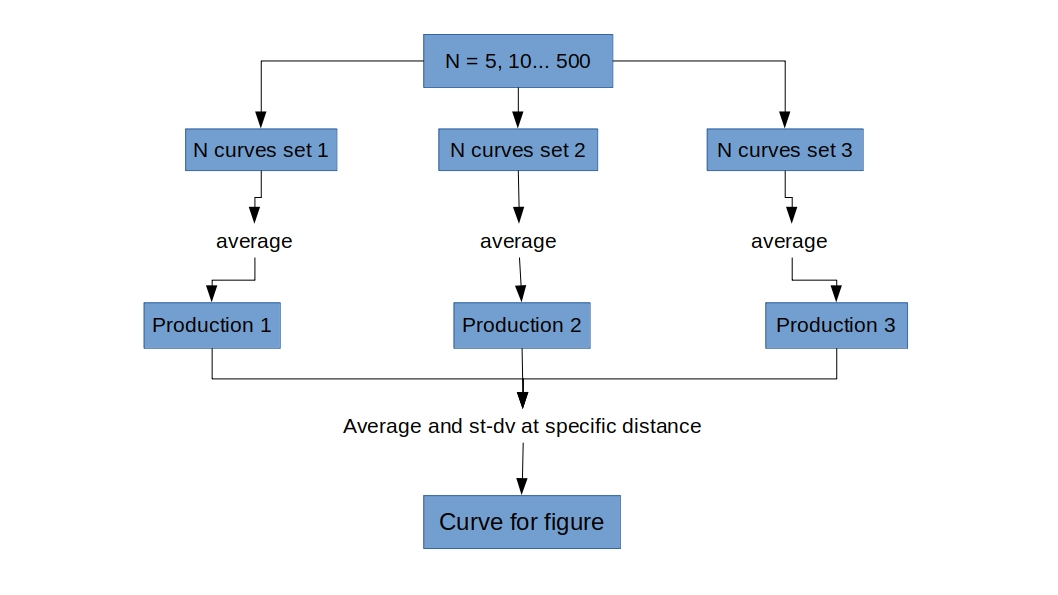
Script follow the next flow chart:
echo 5.calculate_bin.sh
chmod +x 5.calculate_bin.sh
./5.calculate_bin.sh > ./5.calculate_bin.log
Test how the figures are getting.
echo 6.figures.sh
chmod +x 6.figures.sh
./6.figures.sh
Now is time to calculate the deviation using sets of curves.
echo 7.deviation.sh
chmod +x 7.deviation.sh
./7.deviation.sh
One more test to check how is getting the plots based on the outputs of 7.deviation.sh.
chmod +x 8.figures.sh
echo 8.figures.sh
./8.figures.sh
To calculate the virial, we should separate in sets of curves the three contributions [total and vdw (estimated by 7.deviation.sh), and electrostatic (computed with the script in python 9.contributions.py)]. The next script will create all these files in the folder #oligomer#_electrostatic for each virus. Then, the virial will be estimated.
Notes:
- This script just will calculate the virial and electrostatic part based on the best bin and number of curves suitable!
chmod +x 9.virial.sh
echo 9.virial.sh
./9.virial.sh
- This script will create csv, log, and xvg files too.
NOTE: ALL SCRIPTS CAN BE EXECUTE ONCE USING:
chmod +x 0.README
./0.README