Chi-Yun Wu, Jiazhen Rong, Zhang Lab, University of Pennsylvania
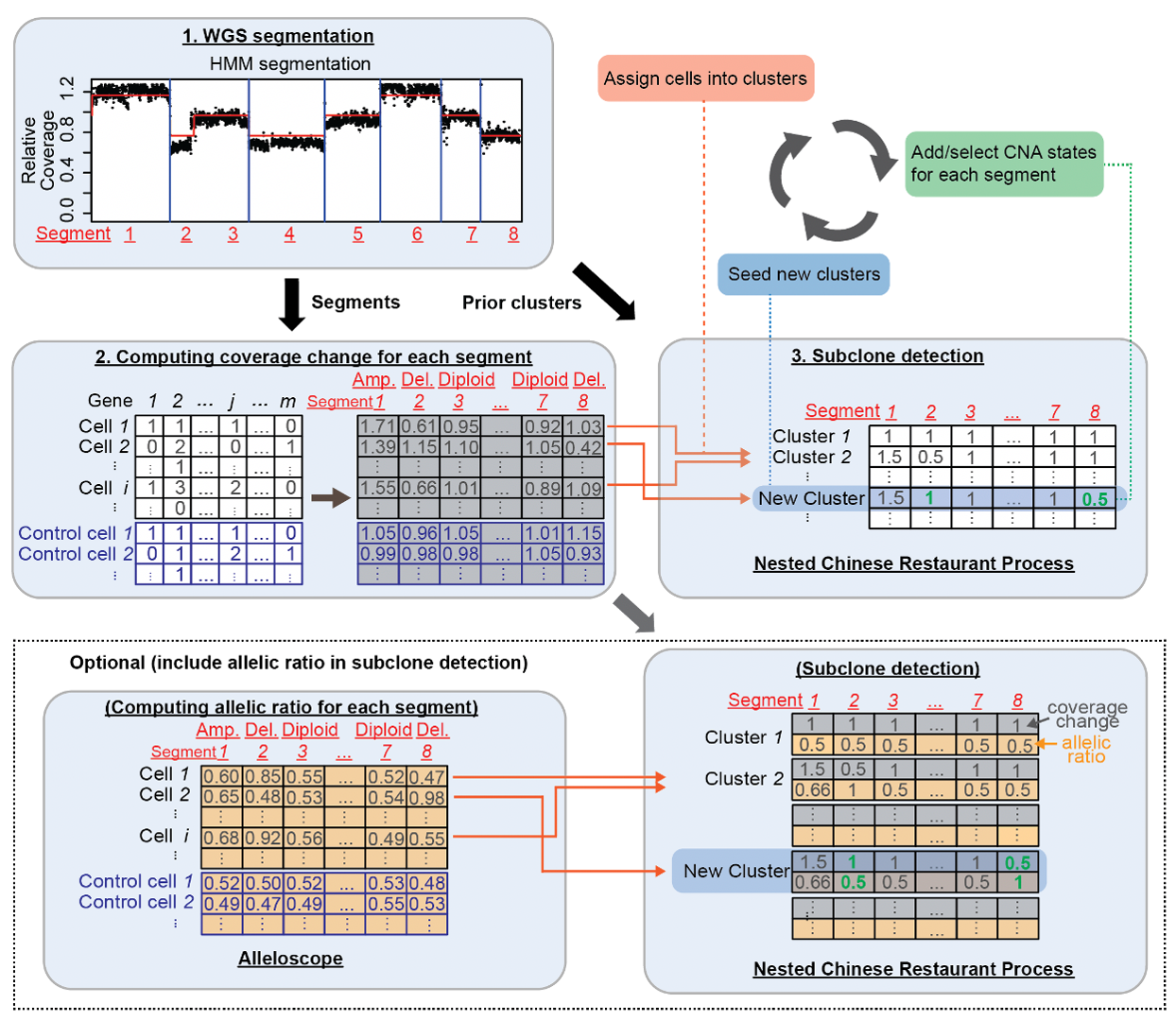
Clonalscope is a subclone detection method based on copy number alterations (CNAs) for single-cell and ST tumor sequencing data. Clonalscope leverages prior information from matched bulk DNA-seq data, and implements a nested Chinese Restaurant Process to model the evolutionary process in tumors. Clonalscope is able to detect subclones, label malignant cells (with the help of matched WGS/WES data), and trace subclones for both scRNA-seq and ST data.
For more information about the method, please check out the manuscript.
- You can install Clonalscope with the code below:
Sys.setenv(R_REMOTES_NO_ERRORS_FROM_WARNINGS="true")
install.packages("devtools")
options(timeout=9999999)
devtools::install_github("seasoncloud/Clonalscope") # install
library(Clonalscope) # load
# It takes <5 mins for the installation.-
You can download example datasets for Clonalscope with the following command:
Using terminal, download the repository.
git clone https://github.com/seasoncloud/Clonalscope.git -
We have included example data in the folder data-raw/.
- Click the links below to read detailed tutorials for different data types.
- scRNA-seq subclone detection and malignant cell labeling
- Spatial Transcriptomic subclone detection
- Spatial Transcriptomic subclone tracing
- scRNA-seq subclone detection and malignant cell labeling without paired WGS/WES
Wu, C.-Y. et al. Cancer subclone detection based on DNA copy number in single cell and spatial omic sequencing data. bioRxiv (2022): https://doi.org/10.1101/2022.07.05.498882