Use this repo to generate vegetation health reports for the input AOI (Area of Interest) geojson or shapefile.
Input AOI --> generates NDVI --> performs kmeans clustersing --> generates PDF report.
- Uses cloud optimized GeoTIFF (COG) format to avoid downloading large files onto your local machine
- Option to provide start date and end date
- Automatically calculates ideal no. of clusters (kmeans) using elbow curve method
- Generates an interactive folium map (html) with identified clusters
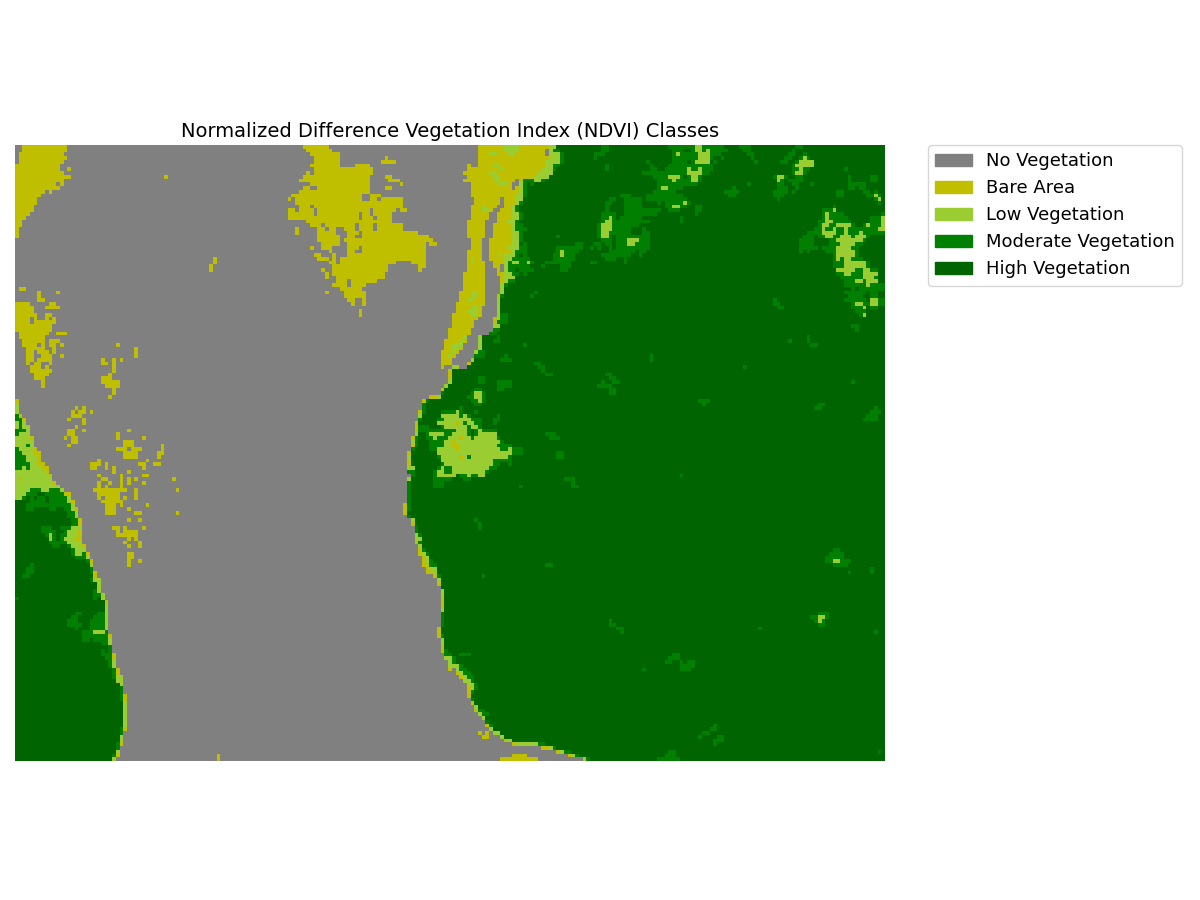
- For each available date, generates a pdf report containing RGB, NDVI, NDVI classes, Kmeans clusters, and clusters superimposed on RGB
- Script to download data can be used as an independent module with options to specify data source (sentinel, landsat, etc.), bands, cloud threshold, etc.
- Applies a precautionary buffer of radius=0 at input AOI, to make sure polygons are valid
Please see Exploratory Data Analysis (EDA) readme for more details on data.
Run the script main.py and pass the following arguments:
- "--aoi": path to aoi vector file (geojson or shapefile)
- "--clusters" [optional]: Number of clusters desired in output. Will auto compute ideal no. of clusters if left blank
- "--start_date" [optioanl]: start date in YYYY-MM-DD format. Will take today's date if left blank
- "--end_date" [optional]: end date in YYYY-MM-DD format. Will take today's date if left blank
- "--out_dir" [optional]:path to directory where data will be generated
- "--crs" [optional]: target CRS of the generated data. If skipped, CRS will match input aoi's CRS
- "--cloud_threshold" [optional]: Cloud cover threshold in %. If skipped, the default value is set to 5
Example 1:python main.py --aoi resources/test_aoi_river.geojson
Example 2:python main.py --aoi resources/test_aoi_river.geojson --start_date 2021-08-17 --clusters 3 --cloud_threshold 15
These steps are for Ubuntu (might differ a bit for MacOS and Windows)
Ensure that GDAL is installed in your environment. See this page for gdal's installation instructions.
Step 1: Clone the repo --> git clone https://github.com/seedlit/vegetation-health-report.git
Step 2: move into the repo --> cd vegetation-health-report
Step 3: Create a new virtual environment --> python3 -m venv venv
Step 4: Activate the virtual environment --> source venv/bin/activate
Step 5: Install the dependencies --> pip install -r requirements.txt
Step 6: Run tests --> python test.py
- remove dependency on gdal. Most of the gdal's functions that I am using can be completed by rastertio or some other library. This would help in creating a pypi package for this repo
- create a package - conda or pypi
- Incorporate the generated folium maps in generated pdf report (as screeshots). Existing solution use selenium to open the html in browser and take a screenshot. But ideally, I would like to avoid that.
- Degub cloud cover checks issue: the cloud cover threshold doesen't seems to be working
- Use multiprocessing to speed up
- Make async. For example, no need to wait for data to get downloaded for all dates. Start wokring as soon as data is downloaded for a date. And remaining downloading can continue in background
- Add area for each NDVI classes. For eg. 70% area belongs to moderate vegetation, etc.