This metagenomics pipeline is mainly divided into two major components. The first component deals with cleaning and removing non-viral contents from the reads and the second with de-novo assembly.
1. Cleaning – removing known higher level organism and bacterial reads
The first step of the cleaning pipeline is to remove the adapters using trim_galore (Krueger). In some cases of metagenomics sample preparation for sequencing ribosomal RNA (rRNA) removal might not be removed before sequencing the sample. In such cases and also in cases it is advisable to remove the rRNAs in silico using RiboPicker (Schmieder et al., 2012). After this, DIAMOND (Buchfink et al., 2014) can be run for each read against the refseq protein database for each file. Krona charts (Ondov et al., 2011) are generated for each DIAMOND output file that describes the read based classification of the sample. The DIAMOND results are then converted to a BLAST tabular output and the Genbank Identifier (GI) column from the output is extracted. GIs are mapped back to NCBI taxonomy databases to extract the corresponding taxonomy and division ID. In our case, we are only interested in the sequences that match viruses, environmental sequences and the sequences that do not match anything in the database. Therefore, any read that is matching protein sequences originating from bacteria, invertebrates, mammals, rodents, phages, plants, vertebrates, primates and synthetic constructs are identified based on the division ID and are filtered from the sample files. Finally, the sample files are properly paired using Prinseq-lite.pl (Schmieder et al., 2011). These reads can then be submitted to the next stage of the pipeline.
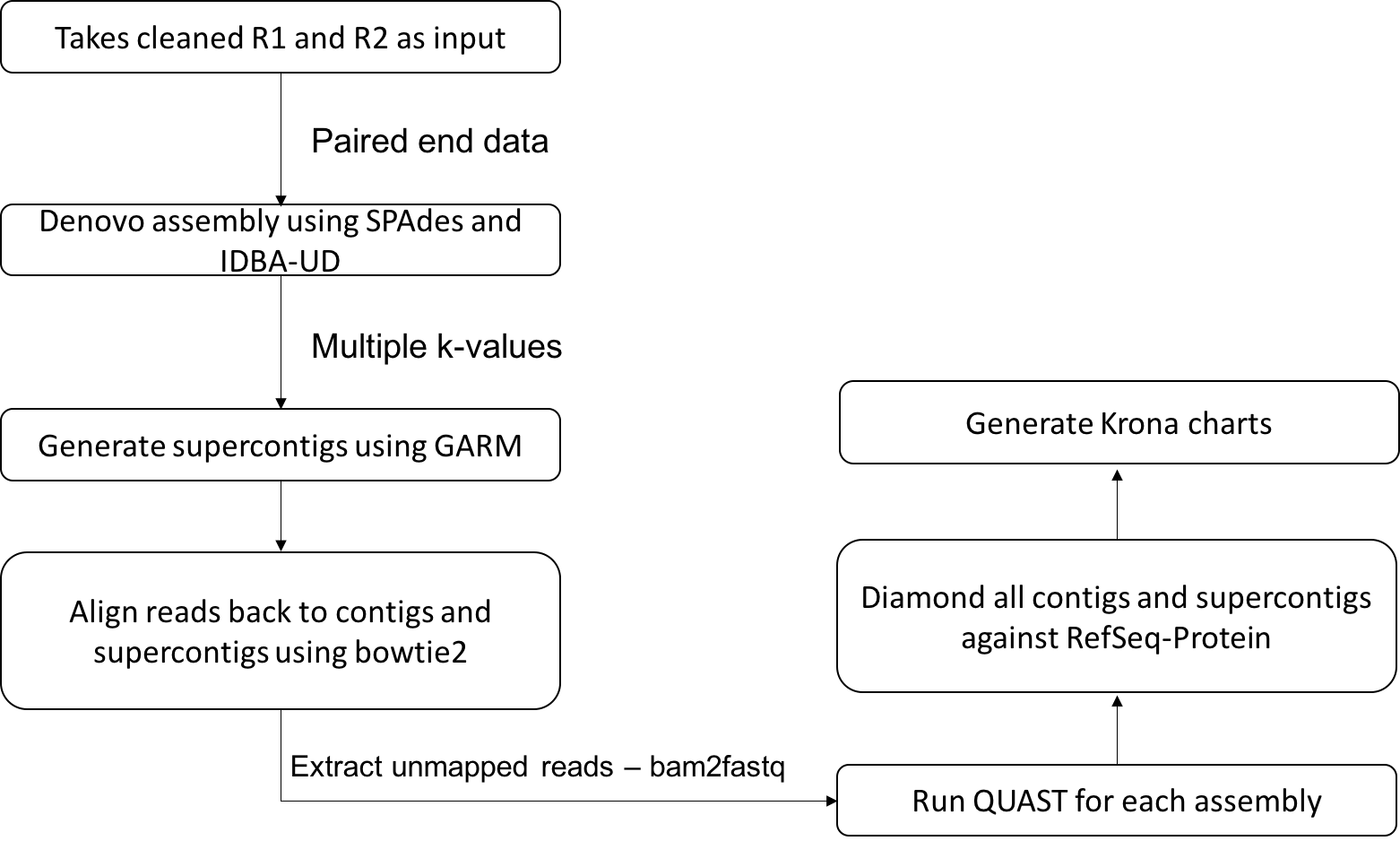
2. De novo assembly
Sequence reads from the previous step of the pipeline can be submitted to this script that carries out the de-novo assembly using SPAdes (Bankevich et al.; Nurk et al., 2013) and IDBA-UD (Peng et al., 2012) with the k-mer values 31,55,77,99 for SPAdes and a range of the k-mer values starting with minimum k of 31 to maximum k of 99 for IDBA-UD.
The assembled contigs from two assembly tools are then merged using GARM (Soto-Jimenez et al., 2014), an assembly merging pipeline that uses mummer to find overlaps between two assemblies and joins them. Reads are aligned back to the contigs (larger than 200) and supercontigs using bowtie2 (Langmead and Salzberg, 2012) and unmapped reads are extracted using bam2fastq. In order to check the assembly quality, a QUAST (Gurevich et al., 2013) analysis is performed for each contig assembly and for supercontigs. Contigs longer than length of 200 and supercontigs generated by GARM are merged and classified using DIAMOND against refseq protein database. Kronatools are used to create interactive HTML output to visualise the BLAST tabular output formatted results generated by DIAMOND.
This pipeline has been applied to field samples of midges from the Scotland, fecal samples of the bats from Brazil, insects from Antarctica.
If you use MetaViC in your project please cite: Modha, S.; Hughes, J.; Bianco, G.; Ferguson, H.M.; Helm, B.; Tong, L.; Wilkie, G.S.; Kohl, A.; Schnettler, E. Metaviromics Reveals Unknown Viral Diversity in the Biting Midge Culicoides impunctatus. Viruses 2019, 11, 865. DOI: https://doi.org/10.3390/v11090865
-
Dennis, Tristan Philip Wesley, William Marciel de Souza, Marc\’\ilio Jorge Fumagalli, Jansen de Araujo, Felipe Gonçalves Motta Maia, Gustavo Olszanski Acrani, Adriano de Oliveira Torres Carrasco, et al. 2018. “Whole-Genome Sequencing of Parvoviruses from Wild and Domestic Animals in Brazil Provides New Insights into Parvovirus Distribution and Diversity.” Viruses. MDPI.
-
Souza, W M, G O Acrani, A Carrasco, M J Fumagalli, M F Romeiro, S Modha, J Hughes, M R T Nunes, P R Murcia, and L T M Figueiredo. 2018. “A58 Identification of Novel Viruses in the Families Flaviviridae (Jigmenvirus), Chuviridae, and Bunyaviridae (Phlebovirus-like) in Ticks from the South of Brazil.” Virus Evolution 4 (suppl_1). Oxford University Press:vey010--057.
-
Souza, William Marciel de, Marilia Farignoli Romeiro, Gilberto Sabino-Santos, Felipe Gonçalves Motta Maia, Marcilio Jorge Fumagalli, Sejal Modha, Marcio Roberto Teixeira Nunes, Pablo Ramiro Murcia, and Luiz Tadeu Moraes Figueiredo. 2018. “Novel Orthohepeviruses in Wild Rodents from São Paulo State, Brazil.” Virology 519. Academic Press:12–16.
-
Souza, William de, Tristan Dennis, Marcílio Fumagalli, Jansen Araujo, Gilberto Sabino-Santos, Felipe Maia, Gustavo Acrani, et al. 2018. “Novel Parvoviruses from Wild and Domestic Animals in Brazil Provide New Insights into Parvovirus Distribution and Diversity.” Viruses 10 (4). Multidisciplinary Digital Publishing Institute:143. https://doi.org/10.3390/v10040143.
-
Marciel de Souza, William, Marcílio Jorge Fumagalli, Jansen de Araujo, Gilberto Sabino-Santos Jr, Felipe Gonçalves Motta Maia, Marilia Farignoli Romeiro, Sejal Modha, et al. 2017. “Discovery of Novel Anelloviruses in Small Mammals Expands the Host Range and Diversity of the Anelloviridae.” https://doi.org/10.1016/j.virol.2017.11.001.
-
Souza, William Marciel de, Marilia Farignoli Romeiro, Marcílio Jorge Fumagalli, Sejal Modha, Jansen de Araujo, Luzia Helena Queiroz, Edison Luiz Durigon, Luiz Tadeu Moraes Figueiredo, Pablo Ramiro Murcia, and Robert James Gifford. 2017. “Chapparvoviruses Occur in at Least Three Vertebrate Classes and Have a Broad Biogeographic Distribution.” Journal of General Virology 98 (2):225–29. https://doi.org/10.1099/jgv.0.000671.
Bankevich,A. et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing.
Buchfink,B. et al. (2014) Fast and sensitive protein alignment using DIAMOND. Nat. Methods, 12, 59–60.
Gurevich,A. et al. (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics, 29, 1072–1075.
Krueger,F. Trim Galore - Babraham Bioinformatics.
Langmead,B. and Salzberg,S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359.
Nurk,S. et al. (2013) Assembling Genomes and Mini-metagenomes from Highly Chimeric Reads. Springer Berlin Heidelberg, pp. 158–170.
Ondov,B.D. et al. (2011) Interactive metagenomic visualization in a Web browser. BMC Bioinformatics, 12, 385.
Peng,Y. et al. (2012) IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinforma. Orig. Pap., 28, 1420–142810.
Schmieder,R. et al. (2012) Identification and removal of ribosomal RNA sequences from metatranscriptomes. Bioinformatics, 28, 433–435.
Schmieder,R. et al. (2011) Quality control and preprocessing of metagenomic datasets. Bioinforma. Appl. NOTE, 27, 863–86410.
Soto-Jimenez,L.M. et al. (2014) GARM: genome assembly, reconciliation and merging pipeline. Curr. Top. Med. Chem., 14, 418–24.
| Tool | Availability |
|---|---|
| Bowtie2 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| DIAMOND | https://github.com/bbuchfink/diamond |
| filter_fastq.pl | https://github.com/lmrodriguezr/enveomics/blob/master/Scripts/FastQ.filter.pl |
| GARM | http://garm-meta-assem.sourceforge.net/ |
| IDBA-UD | https://github.com/loneknightpy/idba |
| Kronatools | https://github.com/marbl/Krona/wiki |
| prinseq-lite.pl | https://sourceforge.net/projects/prinseq/files/ |
| QUAST | http://bioinf.spbau.ru/quast |
| riboPicker | https://sourceforge.net/projects/ribopicker/files/ |
| SPAdes | http://bioinf.spbau.ru/spades |
| Trim Galore | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| weeSAMv1.1 | https://github.com/josephhughes/Sequence-manipulation/blob/master/weeSAMv1.1 |