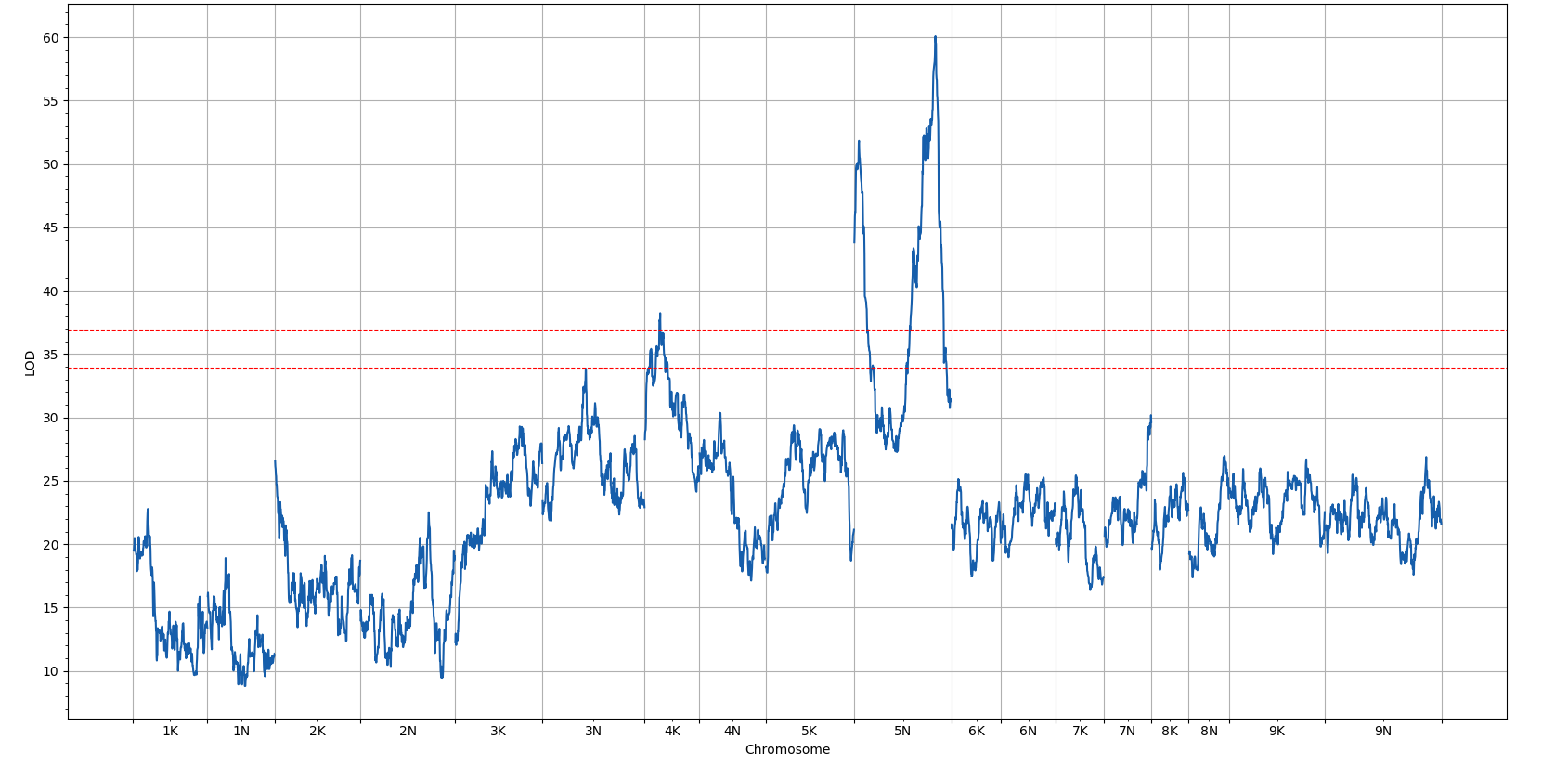
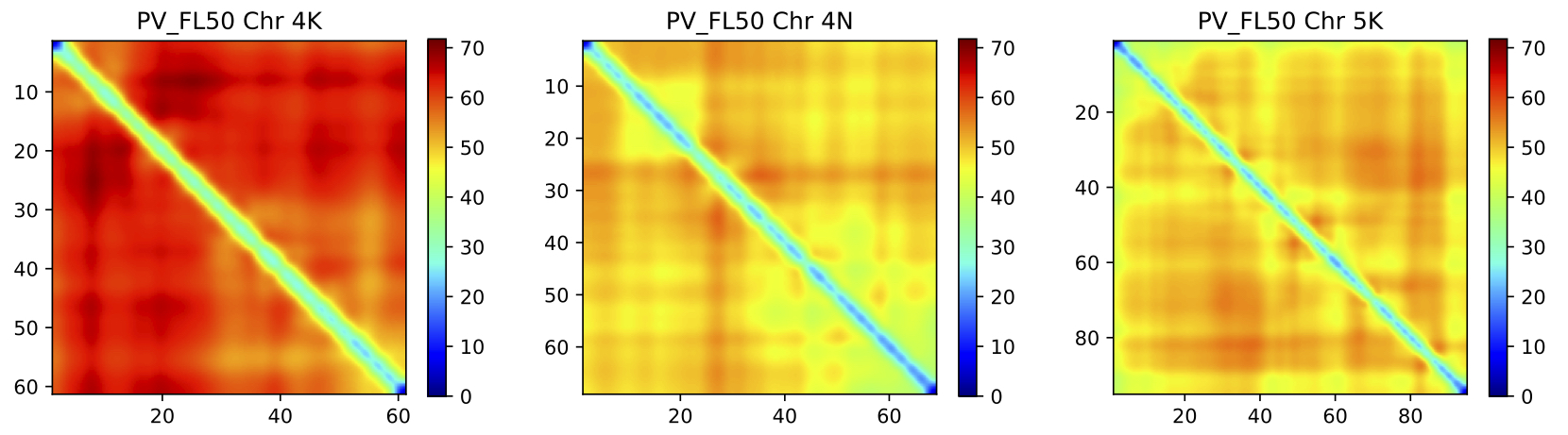
FlxQTL.jl is a a package for a multivariate linear mixed model based QTL analysis tool that supports incorporating information from trait covariates such as time or different environments. The package supports computation of one-dimensional and two-dimensional multivariate genome scans, visualization of genome scans, support for LOCO (leave-one-chromosome-out), computation of kinship matrices, and support for distributed computing.
The package is written in Julia and includes extensive documentation. If you are new to Julia you may want to learn more by looking at Julia documentation. Example data sets are located in the data directory. For details about the method, you may want to read our paper available as a preprint.
Flexible multivariate linear mixed models for structured multiple
traits
Hyeonju Kim, Gregory Farage, John T. Lovell, John K. Mckay, Thomas
E. Juenger, Śaunak Sen
doi: https://doi.org/10.1101/2020.03.27.012690
The package can installed in following ways.
In a Julia REPL, press ] to enter a package mode,
julia> ]
pkg> add FlxQTLOr, equivalently,
julia> using Pkg; Pkg.add("FlxQTL")For installing from the source,
pkg> add https://github.com/senresearch/FlxQTL.jlor,
julia> Pkg.add(url="https://github.com/senresearch/FlxQTL.jl")Currently Julia 1.5 supports for the package.
To remove the package from the Julia REPL,
julia> ]
pkg> rm FlxQTLEquivalently,
julia> using Pkg; Pkg.rm("FlxQTL")The package can be run with OpenBLAS (built-in Julia dense linear
algebra routines) or MKL (Intel's Math Kernel Library). MKL.jl
works best on Intel hardware, although without Intel hardware, it can
improve performance slightly. For installation, and details
see: MKL.jl.