By Chaoran Cheng, Oct 1, 2023
Official implementation of the NeurIPS 23 spotlight paper Equivariant Neural Operator Learning with Graphon Convolution for modeling operators on continuous data.
UPDATE: The pretrained model is available here.
All codes are run with Python 3.9.15 and CUDA 11.6. Similar environment should also work, as this project does not rely
on some rapidly changing packages. Other required packages are listed in requirements.txt.
The QM9 dataset contains 133885 small molecules consisting of C, H, O, N, and F. The QM9 electron density dataset was built by Jørgensen et al. (paper) and was publicly available via Figshare. Each tarball needs to be extracted, but the inner lz4 compression should be kept. We provided code to read the compressed lz4 file.
The Cubic dataset contains electron charge density for 16421 (after filtering) cubic crystal system cells. The dataset was built by Wang et al. (paper) and was publicly available via Figshare. Each tarball needs to be extracted, but the inner xz compression should be kept. We provided code to read the compressed xz file.
WARNING: A considerable proportion of the samples uses the rhombohedral lattice system (i.e., primitive rhomhedral
cell instead of unit cubic cell). Some visualization tools (including plotly) may not be able to handle this.
The MD dataset contains 6 small molecules (ethanol, benzene, phenol, resorcinol, ethane, malonaldehyde) with different geometries sampled from molecular dynamics (MD). The dataset was curated from here by Bogojeski et al. and here by Brockherde et al. The dataset is publicly available at the Quantum Machine website.
We assume the data is stored in the <data_root>/<mol_name>/<mol_name>_<split>/ directory, where mol_name should be
one of the molecules mentioned above and split should be either train or test. The directory should contain the
following files:
structures.npycontains the coordinates of the atoms.dft_densities.npycontains the voxelized electron charge density data.
This is the format for the latter four molecules (you can safely ignore other files). For the former two
molecules, run python generate_dataset.py to generate the correctly formatted data. You can also specify the data
directory with --root and the output directory with --out.
All MD datasets assume a cubic box with side length of 20 Bohr and 50 grids per side. The densities are store as Fourier coefficients, and we provided code to convert them.
Most hyperparameters are specified in the config files. More parameters in the YAML file is self-explanatory. See this readme for more details on modifying the config files. Free feel to modify the config files to suit your needs or to add new models. The pretrained model together with a sample electron density file is available here.
To train the model, run
python configs/qm9.yml --savename testTo evaluate the model, run
python configs/qm9.yml --savename test --mode inf --resume <model_path>To see the visualization of the predicted density, run inference.ipynb with JupyterLab or Jupyter Notebook.
To utilize the code for other (GNN-based) models, you need to register the model class in using
the models.register_model decorator. Your model's forward function should take same arguments as our InfGCN, but the
initialization arguments can be different (see the instructions on modifying the config file).
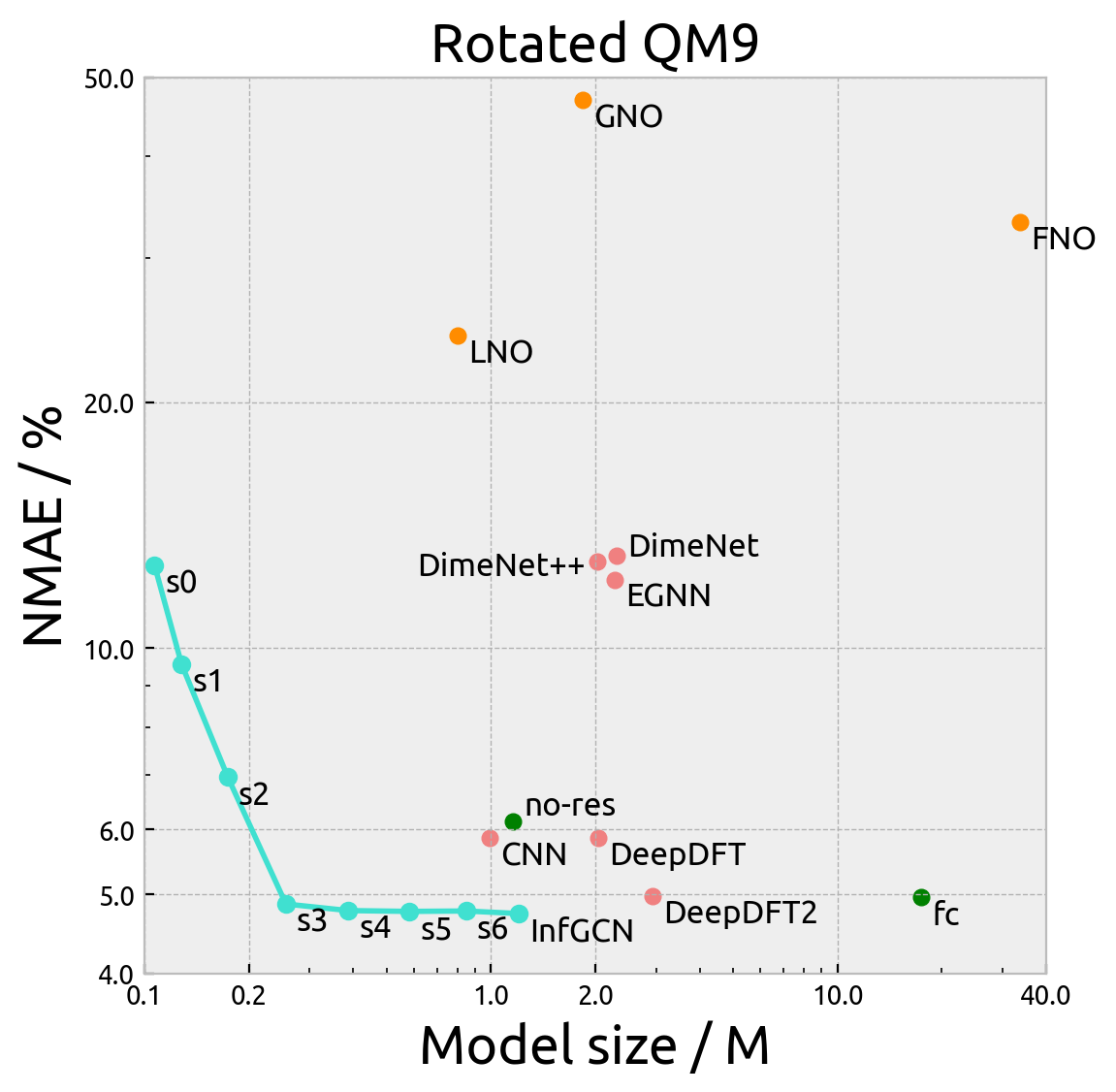
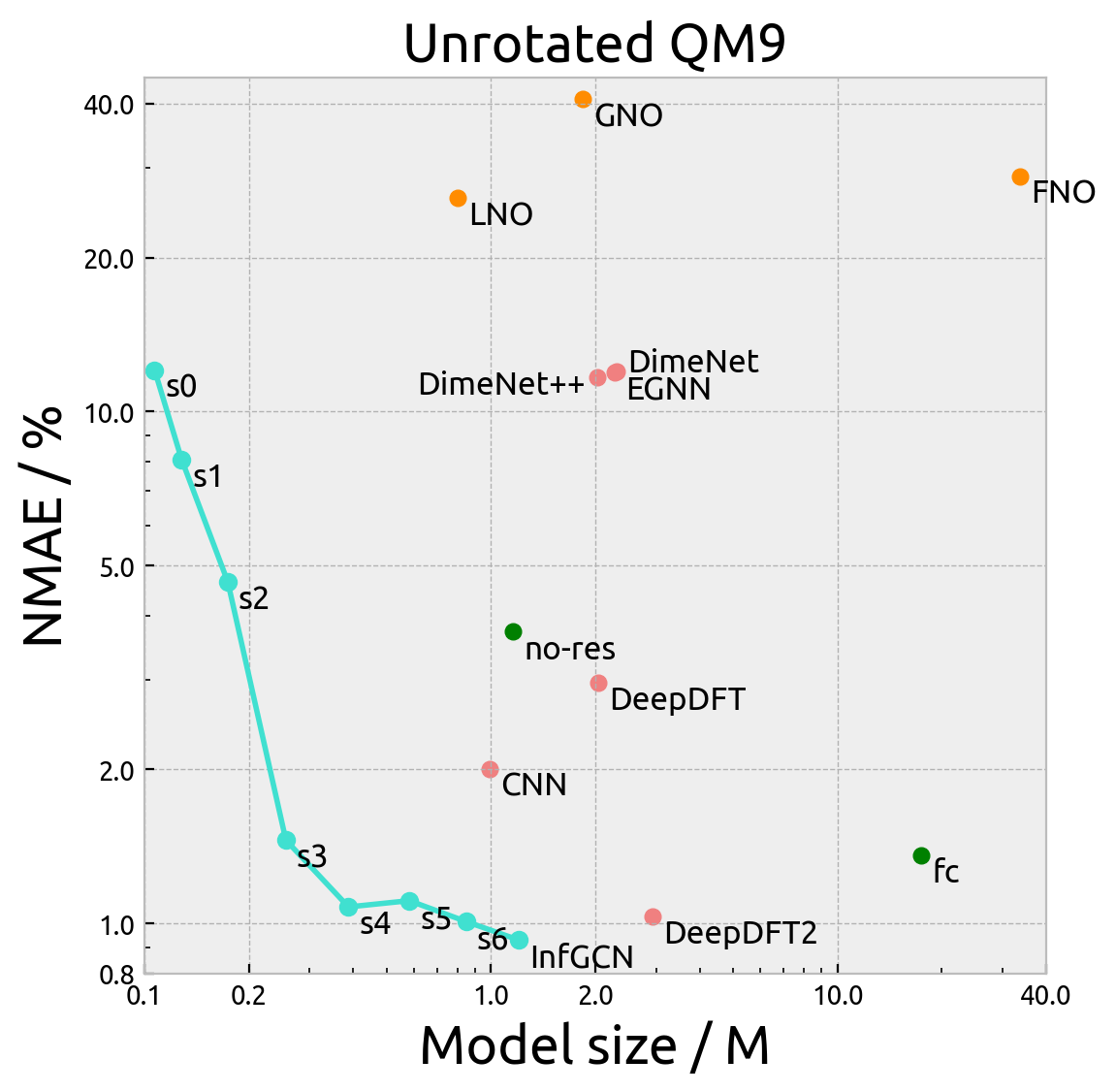
The below figures demonstrate the normalized mean absolute error (NMAE) vs the model size of our model and all the
baseline model on the QM9 dataset. Here, s0 to s6 refer to the maximum degree of spherical harmonics used in the
model (InfGCN is s7). no-res refers to the model without residual connection and fc refers to the model without
fully-connected tensor product. The pink points are interpolation GNNs and oranges points are neural operators.
If you find this code useful, please cite our paper
@InProceedings{Cheng2023infgcn,
title={Equivariant Neural Operator Learning with Graphon Convolution},
author={Chaoran Cheng and Jian Peng},
booktitle={Advances in Neural Information Processing Systems 37: Annual Conference on Neural Information Processing Systems 2023, NeurIPS 2023, December 10-16, 2023},
month={December},
year={2023},
}