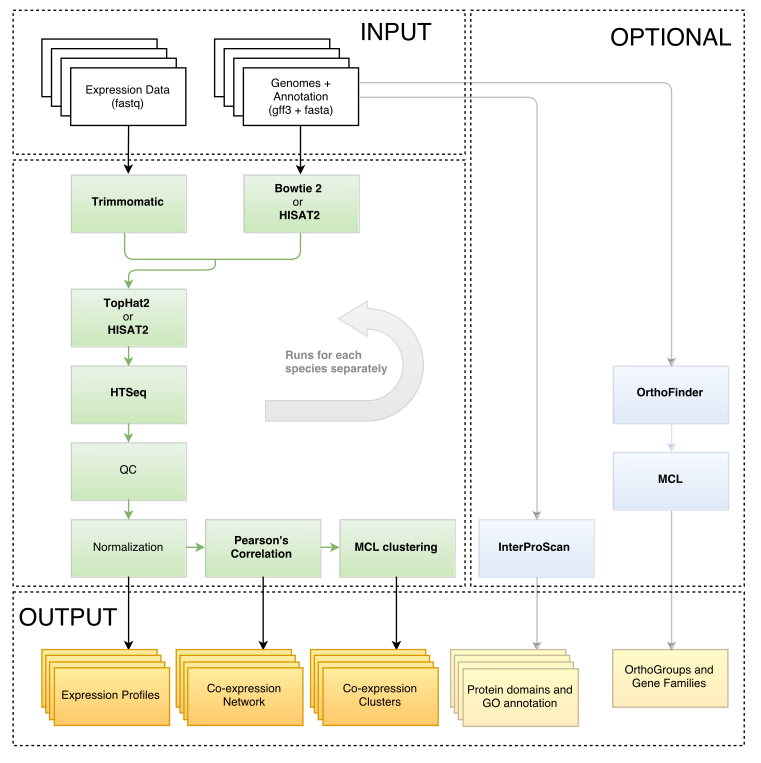
LSTrAP, short for Large Scale Transcriptome Analysis Pipeline, greatly facilitates the construction of co-expression networks from RNA-Seq data. The various tools involved are seamlessly connected and CPU-intensive steps are submitted to a computer cluster automatically.
For more details see the LSTrAP paper below:
Sebastian Proost, Agnieszka Krawczyk and Marek Mutwil (2017) LSTrAP: efficiently combining RNA sequencing data into co-expression networks BMC Bioinformatics 18:444 https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-017-1861-z
- Support for PBS / Torque scheduler (note proper configuration is required)
- HISAT2 can be used as an alternative to BowTie2 and TopHat 2
- Added helper script to do PCA on samples
- Parameter names in data.ini changed, additional options added to config.ini. Check the configuration and update the files accordingly.
LSTrAP wraps multiple existing tools into a single workflow. To use LSTrAP the following tools need to be installed
Steps in bold are submitted to a cluster. Optional steps can be enabled by adding the flag ‑‑enable‑interpro and/or ‑‑enable‑orthology.
Before installing make sure your system meets all requirements. A detailed list of supported systems and required software can be found here.
Use git to obtain a copy of the LSTrAP code
git clone https://github.com/sepro/LSTrAP
Next, move into the directory and copy config.template.ini and data.template.ini
cd LSTrAP
cp config.template.ini config.ini
cp data.template.ini data.ini
Configure config.ini and data.ini using these guidelines
Before running LSTrAP make sure you have all required data. RNA-Seq data needs to be de-multiplexed and de-barcoded, one file per samples and paired-end files need to be named properly (e.g. sample_one_1.fastq.gz and sample_one_2.fastq.gz).
Instructions on how to do this are included here
Once properly configured for your system and data, LSTrAP can be run using a single simple command (that should be executed on the head node).
./run.py config.ini data.ini
Run using HISAT2
./run.py --use-hisat2 config.ini data.ini
Run with InterProScan and/or OrthoFinder
./run.py --enable-orthology --enable-interpro config.ini data.ini
Furthermore, steps can be skipped (to avoid re-running steps unnecessarily). Use the command below for more info.
./run.py -h
- Data preparation
- LSTrAP output
- Quality statistics: How to check the quality of samples and remove problematic samples
- Helper scripts: To acquire data from the Sequence Read Archive and process results.
LSTrAP was developed by Sebastian Proost and Marek Mutwil at the Max-Planck Institute for Molecular Plant Physiology
This work is supported by ERA-CAPS though the EVOREPRO project. The authors would like to thank Andreas Donath for technical support and helpful discussions.
LSTrAP is freely available under the MIT License