27 June 2018 (v0.7.0):
- Back end changed to TensorFlow.
- API/command line options changed slightly. Documentation updated accordingly.
- Updated the default config in ./examples/config/deepmedic with three pathways.
- Refactored/reorganized the code.
28 Oct 2017 (v0.6.1):
- Supporting Theano 0.10beta (v0.9 was found unstable with some cuDnn versions).
- Refactored/reorganized the modules.
- Updated installation process in README, requiring >= theano-0.10.
26 Mar 2017 (v0.6):
- Functionality for transfering parameters and finetuning. See Sec 3.2 below.
- Code refactored to facilitate upcoming extension.
- Important: "Optimization" related configuration parameters moved from modelConfig.cfg to trainConfig.cfg. To reuse old config files, please adjust them to the new format. This should be easy, as all parameters that were moved are under the "Optimization" tagged block of the new trainConfig.cfg.
14 Nov 2016 (v0.5.4):
- Original configuration moved to deepMedicOriginal. Updated config now in deepMedic.
- More memory efficient testing. CNN code has been refactored. Minor fixes.
10 Oct 2016 (v0.5.3):
- Sampling refactored. Now possible to use weighted-maps to sample each class.
4 Aug 2016 (v0.5.2):
- Code in Layer-classes cleaned/commented. Residual Connections enabled. Possible to specify kernel size at FC1.
14 July 2016 (v0.5.1):
- Master branch was updated with better monitoring of training progress and a better plotting script. This version is not backwards compatible. CPickle will fail loading previously trained models from previous versions of the code.
- Previous version of master branch tagged as v0.5. Use this if you wish to continue working with previously trained models.
This project aims to offer easy access to Deep Learning for segmentation of structures of interest in biomedical 3D scans. It is a system that allows the easy creation of a 3D Convolutional Neural Network, which can be trained to detect and segment structures if corresponding ground truth labels are provided for training. The system processes NIFTI images, making its use straightforward for many biomedical tasks.
This document describes how to install and run the software. Accompanying data are provided to run the preset examples and make sure that the system is functioning on your system. This document also describes the main functionality, in order for the user to understand the main processing cycle. For greater details please consult [1]. We hope this project will serve well in making the state-of-the-art Convolutional Networks more accessible in the field of medical imaging.
The system was initially developed for the segmentation of brain lesions in MRI scans. It was employed for our research presented in [1],[2], where a 3D network architecture with two convolutional pathways was presented for the efficient multi-scale processing of multi-modal MRI volumes. If the use of the software positively influences your endeavours, please cite [1].
[1] Konstantinos Kamnitsas, Christian Ledig, Virginia F.J. Newcombe, Joanna P. Simpson, Andrew D. Kane, David K. Menon, Daniel Rueckert, and Ben Glocker, “Efficient Multi-Scale 3D CNN with Fully Connected CRF for Accurate Brain Lesion Segmentation”, Medical Image Analysis, 2016.
[2] Konstantinos Kamnitsas, Liang Chen, Christian Ledig, Daniel Rueckert, and Ben Glocker, “Multi-Scale 3D CNNs for segmentation of brain Lesions in multi-modal MRI”, in proceeding of ISLES challenge, MICCAI 2015.
- 1. Installation and Requirements
- 2. Running the Software
- 3. How it works
- 4. How to run DeepMedic on your data
- 5. Concluding
- 6. Licenses
The system is written in python. The following libraries are required:
- TensorFlow: This is the Deep Learning library that the back end is implemented with.
- NiBabel: The library used for loading NIFTI files.
- Parallel Python: Parallelization library. Note issue#58 when installing it for python3.
- numpy : General purpose array-processing package.
- six : Python compatibility library.
(The below are for unix systems, but similar steps should be sufficient for Windows.)
The software cloned with:
git clone https://github.com/Kamnitsask/deepmedic/
After cloning it, all dependencies can be installed as described below.
If you do not have sudo/root privileges on a system, we suggest you install using a virtual environment. From a bash shell, create a virtual environment in a folder that you wish:
virtualenv -p python2 FOLDER_FOR_ENVS/ve_tf_dmtf # or python3
source FOLDER_FOR_ENVS/ve_tf_dmtf/bin/activate # If using csh, source ve_tf_dmtf/bin/activate.csh
Then continue with the steps below.
Install TensorFlow (TF): Please follow instructions on (https://www.tensorflow.org/install/). By consulting the previous link, ensure that your system has CUDA version and cuDNN versions compatible with the tensorflow version you are installing.
$ pip install --upgrade tensorflow-gpu # or plain tensorflow, if you dont have a capable GPU.2
Problem installing TF on MacOS: Seems there is often a problem installing TF on MacOS.
If the above fails, eg giving a Could not find version that satisfies requirement tensorflow, see the appropriate documentation for MacOS of TF in the above link.
Something I've found working on Macs installing TF with the following:
python2 pip install --upgrade https://storage.googleapis.com/tensorflow/mac/cpu/tensorflow-1.8.0-py2-none-any.whl # adapt python2, py2, or version of TF as required.
Install DeepMedic and rest of its dependencies:
$ cd DEEPMEDIC_ROOT_FOLDER
$ pip install .
This will grab rest of dependencies described in Sec.1. Note: If you are using python3, please see issue#58. The pp module will need a manual (easy) installation of a specific version, that is compatible with python3.
Small networks can be run on the cpu. But 3D CNNs of considerable size require processing on the GPU. For this, an installation of Nvidia’s CUDA is n eeded. Make sure to acquire a version compatible with your GPU drivers. TensorFlow needs to be able to find CUDA’s compiler, the nvcc, in the environment’s path. It also dynamically links to c ublas.so libraries, which need to be visible in the environment’s.
Prior to running DeepMedic on the GPU, you must manually add the paths to the folders containing these files in your environment's variables. As an example in a bash shell:
$ export CUDA_HOME=/path/to/cuda # If using cshell instead of bash: setenv CUDA_HOME /path/to/cuda
$ export LD_LIBRARY_PATH=/path/to/cuda/lib64
$ export PATH=/path/to/cuda/bin:$PATH
We highly suggest you also install cuDNN (link to instructions). It is easy (simply copy cuDNN files in your Cuda folders in unix) and really offers high acceleration (eg a full MR brain volume segmentation in under 20 secs, in comparison to 1.40 mins without).
-
DeepMedic processes NIFTI files only. All data should be in the .nii format.
-
The input modalities, ground-truth labels, ROI masks and other images of each subject need to be co-registered (per-subject, no need for inter-subject registration).
-
The images of each subject should have the same dimensions (per subject, no need for whole database). This is, the number of voxels per dimension must be the same for all images of a subject.
-
Resample all images in the database to the same voxel size. The latter is needed because the kernels (filters) of the DeepMedic need to correspond to the same real-size patterns (structures) for all subjects.
-
Make sure that the ground-truth labels for training and evaluation represent the background with zero. The system also assumes that the task’s classes are indexed increasing by one (not 0,10,20 but 0,1,2).
-
You are strongly advised to normalize the intensity of the data within the ROI to a zero-mean, unit-variance space. Our default configuration significantly underperforms if intensities are in another range of values.
Note for large images: Large 3D CNNs are computationally expensive. Consider downsampling the images or reducing the size of the network if you encounter computational difficulties. The default configuration of DeepMedic was applied on scans of size around 200x200x200.
The source code of the DeepMedic is provided in the folder deepmedic. Users should not need to touch this folder. The software comes with a command line interface, deepMedicRun. Running it with the help option:
./deepMedicRun -h
brings up the available actions for the creation, training and testing of CNN models. All actions require a large number of configuration parameters, which are read from configuration files.
In the examples/configFiles folder we provide two sets of configuration files. Firstly, the configuration of a very small network is given in examples/configFiles/tinyCnn/. This network can be trained within minutes on a CPU. It's a simple example to make sure everything works. We also provide the full configuration of the DeepMedic model, as employed in [1], in the folder examples/configFiles/deepMedic/.
The above configuration files are pre-set to point to accompanying .nii files, provided in examples/dataForExamples/. Those NIFTIs serve as input to the networks in our examples. This data are modified versions of images from the Brain Tumor Segmentation challenge (BRATS 2015).
We will here train a tiny CNN model and make sure everything works as expected. Further explanations on the use of the software are provided in the next section.
NOTE: First see Section 1.2 for installation of the required packages.
Lets train a model:
./deepMedicRun -model ./examples/configFiles/tinyCnn/model/modelConfig.cfg \
-train examples/configFiles/tinyCnn/train/trainConfigWithValidation.cfg
This command parses the given model-configuration file that defines the architecture and creates the corresponding CNN model. It then parses the training-config that specifies metaparameters for the training scheme. The folder ./examples/output/ should have been created by the process, where all output is saved. The model will then be trained for two epochs. All output of the process is logged for later reference. This should be found at examples/output/logs/trainSessionWithValidTiny.txt. After each epoch the trained model is saved at examples/output/saved_models/trainSessionWithValidTiny. Tensorflow saves the model in form of checkpoint files. You should find ./examples/output/cnnModels/tinyCnn.initial.DATE+TIME.model.ckpt.[data..., index] created after each epoch. Each set of DATE+TIMEmodel.ckpt[data,index] is refered to as one checkpoint, i.e. a saved model. Finally, after each epoch, the model performs segmentation of the validation images and the segmentation results (.nii files) should appear in examples/output/predictions/trainSessionWithValidTiny/predictions/. If the training finishes normally (should take 5 mins) and you can see the mentioned files in the corresponding folders, beautiful. Briefly rejoice and continue...
You can plot the training progress using an accompanying script, which parses the training logs:
python plotTrainingProgress.py examples/output/logs/trainSessionWithValidTiny.txt -d
Now lets test with the trained model (replace DATE+TIME):
./deepMedicRun -model ./examples/configFiles/tinyCnn/model/modelConfig.cfg \
-test ./examples/configFiles/tinyCnn/test/testConfig.cfg \
-load ./examples/output/saved_models/trainSessionWithValidTiny/tinyCnn.trainSessionWithValidTiny.final.DATE+TIME.model.ckpt
Of course replace DATE+TIME accordingly.
Note that we specify which previously-trained model/checkpoint to load parameters from with the -load option. But please note, the path given does NOT correspond neither to the data nor index file. It rather needs to refer to the checkpoint set (i.e., should end with .model.ckpt). Tensorflow's loader peculiarity. This process should perform segmentation of the testing images and the results should appear in examples/output/predictions/testSessionTiny/ in the predictions folder. In the features folder you should also find some files, which are feature maps from the second layer. DeepMedic gives you this functionality (see testConfig.cfg). If the testing process finishes normally and all output files seem to be there, everything seems to be working! On the CPU...
Now lets check the important part... If using the DeepMedic on the GPU is alright on your system. First, delete the examples/output/ folder for a clean start. Now, most importantly, place the path to CUDA's nvcc into your PATH and to the cublas.so in your LD_LIBRARY_PATH (see section 1.3)
You need to perform the steps we did before for training and testing with a model, but on the GPU. To do this, repeat the previous commands and pass the additional option -dev cuda. For example:
./deepMedicRun -model ./examples/configFiles/tinyCnn/model/modelConfig.cfg \
-train ./examples/configFiles/tinyCnn/train/trainConfigWithValidation.cfg \
-dev cuda0
You can replace 0 to specify another device number, if your machine has multiple GPUs. The processes should result in similar outputs as before. Make sure the process runs on the GPU, by running the command nvidia-smi. You should see your python process assigned to the specified GPU. If all processes finish as normal and you get no errors, amazing. Now it seems that really everything works :) Continue to the next section and find more details about the DeepMedic and how to use the large version of our network!
Possible problems with the GPU: If TensorFlow does not find correct versions for CUDA and cuDNN (depends on TensorFlow version), it will fall back to the CPU version by default. If this happens, right after the model creation and before the main training process starts, some warnings will be thrown by TensorFlow, along the lines below:
2018-06-06 14:39:34.036373: I tensorflow/core/platform/cpu_feature_guard.cc:140] Your CPU supports instructions that this TensorFlow binary was not compiled to use: AVX2 FMA
2018-06-06 14:39:35.676554: E tensorflow/stream_executor/cuda/cuda_driver.cc:406] failed call to cuInit: CUDA_ERROR_NO_DEVICE
2018-06-06 14:39:35.676616: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:158] retrieving CUDA diagnostic information for host: neuralmedic.doc.ic.ac.uk
2018-06-06 14:39:35.676626: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:165] hostname: neuralmedic.doc.ic.ac.uk
2018-06-06 14:39:35.676664: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:189] libcuda reported version is: 384.111.0
2018-06-06 14:39:35.676699: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:193] kernel reported version is: 384.111.0
2018-06-06 14:39:35.676708: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:300] kernel version seems to match DSO: 384.111.0
If the process does not start on the GPU as required, please ensure you have CUDA and cuDNN versions that are compatible with the TF version you have (https://www.tensorflow.org/install), and that you environment variables are correctly setup. See Section 1.4 about some pointers, and the CUDA website.
Previously we briefly discussed how to quickly run a pre-set example with a tiny CNN, just so you can check whether everything works on your system. In this section we will go through the process in a bit more detail. We also explain the main parameters that should be specified in the configuration files, in order for you to tailor the network and process to your needs.
The .cfg configuration files in examples/configFiles/deepMedicOriginal/ display the parameters used in our work in [1]. In an attempt to make it simpler for the user, we also provide a cleaner version of the configuration files, named with "Less", where many parameters are "hidden". They are internally passed values that worked well in our experiments. Finally, the config files in examples/configFiles/deepMedic/ provide a network configuration that we will be gradually updating with components that seem to improve the overall performance of the system.
Note: The config files are parsed as python scripts, thus follow python syntax. Any commented-out configuration variables are internally given default values.
When performing training or testing, we need to define the architecture of the network. For this, we point to a model-config file, using the option -model :
-model ./examples/configFiles/deepMedic/model/modelConfig.cfg
After reading the parameters given in modelConfig.cfg, the graph of a network is created internally. The session prints all parameters that are used for the model-creation on the screen and to the log.txt file.
Parameters for defining Network Architecture
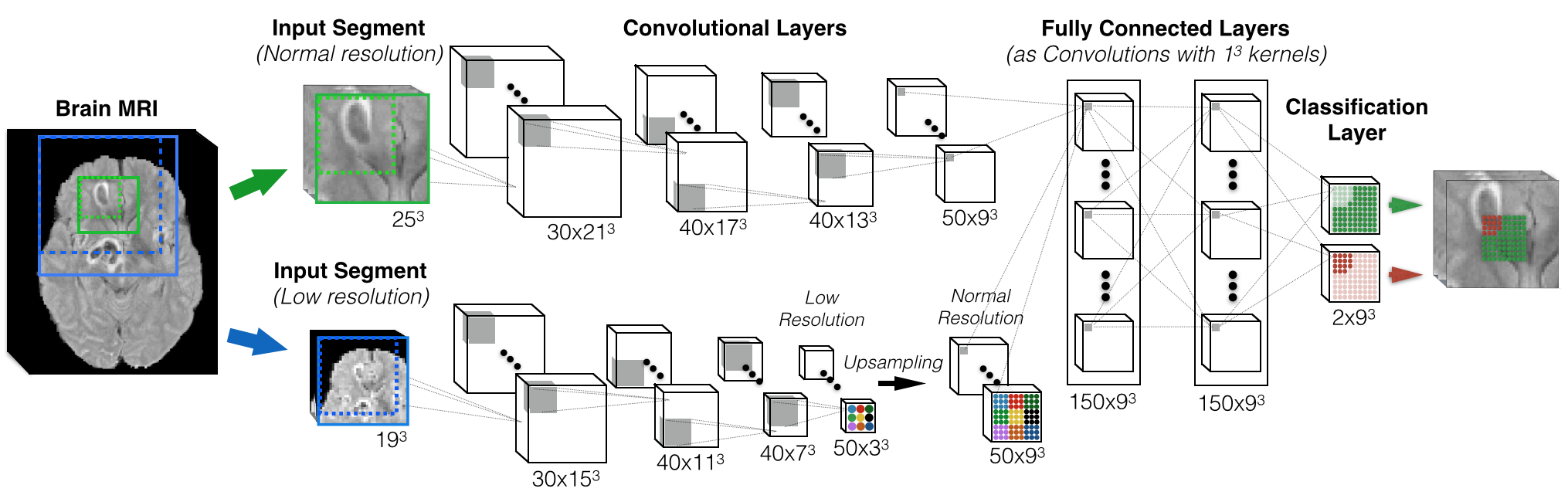 Figure 1: An example of a double-pathway architecture for multi-scale processing. At each layer, the number and size of feature maps (FMs) is depicted in the format (Number-Of-FMs x Dimensions). Actual DeepMedic has 11 layers by default.
Figure 1: An example of a double-pathway architecture for multi-scale processing. At each layer, the number and size of feature maps (FMs) is depicted in the format (Number-Of-FMs x Dimensions). Actual DeepMedic has 11 layers by default.
The main parameters to specify the CNN model are the following.
Generic:
- modelName: This is used for naming the checkpoints when saving every epoch of training. Change it to distinguish between architectures.
- folderForOutput: The main output folder. Saved model and logs will be placed here.
Task Specific:
- numberOfOutputClasses: DeepMedic is multiclass system. This number should include the background, and defines the number of FMs in the last, classification layer (=2 in Fig.1)
- numberOfInputChannels: Specify the number of modalities/sequences/channels of the scans.
Architecture:
- numberFMsPerLayerNormal: A list which needs to have as many entries as the number of layers in the normal pathway that we want to create. Each entry is a number, which defines the number of feature-maps in the corresponding layer ([30, 40, 40, 50] in fig1)
- kernelDimPerLayerNormal: The dimensions of the kernels per layer. ([[5,5,5], [5,5,5], [5,5,5], [5,5,5]] in Fig.1.)
- useSubsampledPathway: Setting this to “True” creates a subsampled-pathway, with the same architecture as the normal one. “False” for single-scale processing with the normal pathway only. Additional parameters allow tailoring this pathway further.
- numberFMsPerLayerFC: The final layers of the two pathways are contatenated. This parameter allows the addition of extra hidden FC layers before the classification layer. The number of entries specified how many extra layers, the number of each entry specifies the number of FMs in each layer. Final classification layer not included ([[150], [150]] in Fig.1).
Image Segments and Batch Sizes:
- segmentsDim(Train/Val/Inference): The dimensions of the input-segment. Different sizes can be used for training, validation, inference (testing). Bigger sizes require more memory and computation. Training segment size greatly influences distribution of training samples ([25,25,25] in Fig.1). Validation segments are by default as large as the receptive field (one patch). Size of testing-segments only influences speed.
- Batch Size : The number of segments to process simultaneously on GPU. In training, bigger batch sizes achieve better convergence and results, but require more computation and memory. Batch sizes for Validation and Inference are less important, greater once just speedup the process.
More variables are available, but are of less importance (regularization, optimizer, etc). They are described in the config files of the provided examples.
You train a model using your manually segmented, ground-truth annotations. For this, you need to point to a configuration file with the parameters of the training session. We use the -model option, that defines a network architecture, together with the -train option, that points to a training-config files, that specifies parameters about the training scheme and the optimization:
./deepMedicRun -model ./examples/configFiles/deepMedic/model/modelConfig.cfg \
-train ./examples/configFiles/deepMedic/train/trainConfig.cfg \
-dev cuda0
Note that you can change 0 with another number of a GPU device, if your machine has multiple GPUs.
The Training Session
During a training session, the following cycle is followed:
For each epoch {
For each subepoch {
Load Validation Cases
Extract Validation Segments
Perform Validation-On-Samples (in batches)
Load Training Cases
Extract Training Segments
Perform Training (in batches)
Report Accuracy over subepoch’s samples (Val/Train).
}
Report Accuracy over whole epoch (Val/Train)
Lower Learning Rate if required.
Save the model’s state
Perform Full-Inference on Validation Cases (every few epochs)
Report DSC of full inference
Save predictions from Full-Inference (segm./prob.maps/features)
}
The validation on samples and the full segmention of the scans of validation subjects are optional.
Plotting Training Progress
The progress of training can be plotted by using the accompanying plotTrainingProgress.py script, which parses the training logs for the reported validation and training accuracy metrics. A common usage example is:
python plotTrainingProgress.py examples/output/logs/trainSession\_1.txt examples/output/logs/trainSession\_2.txt \
-d -m 20 -c 1
Try option -h for help. Here, two logs/experiments are specified, to plot metrics for both to compare. Any number is allowed. -d requests a detailed plot with more metrics. -m 20 runs a moving average over 20 subepochs for smoothing the curves. -c 1 requests plotting class with label=1. Note that in case of multiple labels, -c 0 actually reports the metrics NOT for the background class (as we did not find this useful in most applications), but rather for the whole-foreground class, which can be imagined as if all labels except 0 (assumed background) are fused into one.
Resuming an Interrupted Training Session
A training session can be interrupted for various reasons. Because of this, the state of the model is saved in the end of each epoch and can be found in the output folder. Except for its trainable kernels, we also save the state of the optimizer and parameters of the training session (eg number of epochs trained, current learning rate) in order to be able to seamlessly continue it. An interrupted training session can be continued similarly to how it was started, but by additionally specifying the saved model checkpoint where to load trained parameters and continue from:
./deepMedicRun -model ./examples/configFiles/deepMedic/model/modelConfig.cfg \
-train ./examples/configFiles/deepMedic/train/trainConfig.cfg \
-load ./examples/output/saved_models/trainSessionDm/deepMedic.trainSessionDm.DATE+TIME.model.ckpt \
-dev cuda0
Alternatively, the checkpoint can be defined in the trainConfig file. Importantly, the path must NOT point to the .index or .data files. It must be ending with the .model.ckpt, so that the loader can then find the matching .index and .data files.
Pre-Trained Models, fine-tuning
Common practice with neural networks is to take a network pre-trained on one task/database, and fine-tune it for a new task by training on a second database. This can be naturally done pointing to the pretrained network's checkcpoint (-load) and its architecture (-model) when starting to train. Very importantly though, one may need to reset the state of the trainer at the beginning of the fine-tuning, secondary session. Without this, the trainer will still have a saved state (number of epochs trained, velocities of momentum, learning rate etc) from the first training session. All these parameters can be reset, so that they are reinitialized by the new training-session configuration simply with the -resetopt option, as follows:
./deepMedicRun -model ./examples/configFiles/deepMedic/model/modelConfig.cfg \
-train ./examples/configFiles/deepMedic/train/trainConfigForRefinement.cfg \
-load ./path/to/pretrained/network/filename.DATE+TIME.model.ckpt \
-resetopt \
-dev cuda0
Training Parameters
Generic Parameters:
- sessionName: The name of the session. Used to save the trained models, logs and results.
- folderForOutput: The main output folder.
- cnnModelFilePath: path to a saved CNN model (in case one wants to resume training. Disregarded if -load is used.).
Input for Training:
- channelsTraining: For each of the input channels, this list should hold one entry. Each entry should be a path to a file. These files should list the paths to the corresponding channels for each of the training subjects. See the pre-set files given in the examples and all these should easily become clear.
- gtLabelsTraining: the path to a file. That file should list the paths to the ground-truth labels for all training subjects.
- roiMasksTraining: In many tasks we can easily define a Region Of Interest and get a mask of it. For instance by excluding the air in a body scan, or take the brain-mask in a brain scan. In this case, this parameter allows pointing to the roi-masks for each training subject. Sampling or inference will not be performed outside this area, focusing the learning capacity of the network inside it. If this is not available, detete or comment this variable out and sampling will be performed on whole volumes.
Training Cycle:
- numberOfEpochs: Total number of epochs until the training finishes.
- numberOfSubepochs: Number of subepochs to run per epoch
- numOfCasesLoadedPerSubepoch: At each subepoch, the images from maximum that many cases are loaded to extract training samples. This is done to allow training on databases that may have hundreds or thousands of images, and loading them all for sample-extraction would be just too expensive.
- numberTrainingSegmentsLoadedOnGpuPerSubep: At every subepoch, we extract in total this many segments, which are loaded on the GPU in order to perform the optimization steps. Number of optimization steps per subepoch is this number divided by the batch-size-training (see model-config). The more segments, the more GPU memory and computation required.
Learning Rate Schedule:
- typeOfLearningRateSchedule : Schedules to lower the Learning Rate with. 'stable' keeps LR constant. 'predef' lowers it at predefined epochs, requiring the user to specify at which epochs to lower LR. Auto lowers LR when validation accuracy plateaus (unstable). 'poly' slowly lowers LR over time. We advice to use constant LR, observe progress of training by plotting it (see above), and lower LR manually when improvement plateaus by creating your own 'predef' schedule. Otherwise, use 'poly', but make sure that training is long enough for convergence, by experimenting a bit with the total number of training epochs.
Data Augmentation:
- reflectImagesPerAxis: Specify whether you d like the images to be randomly reflected in respect to each axis, for augmentation during training.
- performIntAugm: Randomly apply a change to segments’ intensities: I' = (I + shift) * multi
Validation:
- performValidationOnSamplesThroughoutTraining, performFullInferenceOnValidationImagesEveryFewEpochs: Booleans to specify whether we want to perform validation, since it is actually time consuming.
- channelsValidation, gtLabelsValidation, roiMasksValidation: Similar to the corresponding training entries. If default settings for validation-sampling are enabled, sampling for validation is done in a uniform way over the whole volume, to achieve correct distribution of the classes.
- numberValidationSegmentsLoadedOnGpuPerSubep: on how many validation segments (samples) to perform the validation.
- numberOfEpochsBetweenFullInferenceOnValImages: Every how many epochs to perform full-inference validation. It might be slow to process all validation cases often.
- namesForPredictionsPerCaseVal: If full inference is performed, we may as well save the results to visually check progress. Here you need to specify the path to a file. That file should contain a list of names, one for each case, with which to save the results. Simply the names, not paths. Results will be saved in the output folder.
When a training epoch is finished, the model’s state is saved. These models can be used for segmenting previously unseen scans. A testing configuration file has to be specified. Testing can be started in two ways.
a) A model is specified straight from the command line.
./deepMedicRun -model ./examples/configFiles/deepMedic/model/modelConfig.cfg \
-test ./examples/configFiles/deepMedic/test/testConfig.cfg \
-load ./path-to-saved-model/filename.model.ckpt \
-dev cuda0
b) The path to a saved model can be instead specified in the testing config file, and then the -load option can be ommited. Note: A file specified by -load option overrides any specified in the config-file.
After the model is loaded, inference will be performed on the testing subjects. Predicted segmentation masks, posterior probability maps for each class, as well as the feature maps of any layer can be saved. If ground-truth is provided, DeepMedic will also report DSC metrics for its predictions.
Note that this testing procedure is similar to the full-inference procedure performed on validation subjects every few training epochs.
Testing Parameters
Main Parameters:
- sessionName: The name for the session, to use for saving the logs and inference results.
- folderForOutput: The output folder to save logs and results.
- cnnModelFilePath: The path to the cnn model to use. Disregarded if specified from command line.
- channels: List of paths to the files that list the files of channels per testing case. Similar to the corresponding parameter for training.
- namesForPredictionsPerCase: Path to a file that lists the names to use for saving the prediction for each subject.
- roiMasks: If masks for a restricted Region-Of-Interest can be made, inference will only be performed within it. If this parameter is omitted in the config file, whole volume is scanned.
- gtLabels: Path to a file that lists the file-paths to Ground Truth labels per case. Not required for testing, but if given, DSC accuracy metric is reported.
Saving Predictions:
- saveSegmentation, saveProbMapsForEachClass : Specify whether you would like the segmentation masks and the probability maps of a class saved.
Saving Feature Maps:
- saveIndividualFms, saveAllFmsIn4DimImage : Specify whether you would like the feature maps saved. Possible to save each FM in a separate files, or create a 4D file with all of them. Note that FMs are many and the 4D file can be several hundreds of MBs, or GBs.
- minMaxIndicesOfFmsToSaveFromEachLayerOfABCPathway : Because the number of FMs is large, it is possible to specify particular FMs to save. Provide the minimum (inclusive) and maximum (exclusive) index of the FMs of the layers that you would like to save (indexing starts from 0).
In examples/configFiles/deepMedicOriginal/ we provide the configuration of the network as employed in our work in [1], and very similar to the model employed in our winning contribution for the ISLES 2015 challenge [2]. The config files named with “Less” are “cleaner” versions to make them more readable, where many parameters are omitted/hidden (they are passed default values internally). The configuration of these two models is exactly the same. In examples/configFiles/deepMedic/ we provide a configuration which we will be gradually updating with any components we find generally well behaved. You are adviced to use the latter, bearing in mind that behavior might slightly change between version (hopefully for the best!).
To run the DeepMedic on your data, the following are the minimum steps you need to follow:
a) Pre-process your data as described in Sec. 1.4. Do not forget to normalise them to a zero-mean, unit-variance space. Produce ROI masks (for instance brain masks) if possible for the task.
b) In the modelConfig.cfg file, change the variable numberOfOutputClasses = 5 to the number of classes in your task (eg 2 if binary), and numberOfInputChannels = 2 to the number of input modalities. Now you are ready to create the model via the -newModel option.
c) (optional) If you want to train a bigger or smaller network, the easiest way is to increase/decrease the number of Feature Maps per layer. This is done by changing the number of FMs in the variable numberFMsPerLayerNormal = [30, 30, 40, 40, 40, 40, 50, 50].
d) Before you train a network you need to alter the trainConfig.cfg file, in order to let the software know where your input images are. The variable channelsTraining = ["./trainChannels_flair.cfg", "./trainChannels_t1c.cfg"] is pre-set to point to two files, one for each of the input variables. Adjust this for your task.
e) Create your files that correspond to the above ./trainChannels_flair.cfg, trainChannels_t1c.cfg files for your task. Each of these files is essentially a list. Every file has an entry for each of the training subjects. The entry is the path to the .nii file with the corresponding modality image for the subject. A brief look to the provided exemplary files should make things clear.
f) Do the same process in order to point to the ground-truth labels for training via the variable gtLabelsTraining = "./trainGtLabels.cfg" and to ROI masks (if available) via roiMasksTraining = "./trainRoiMasks.cfg".
g) If you wish to periodically perform validation throughout training, similar to the above, point to the files of validation subjects via the variables channelsValidation, gtLabelsValidation and roiMasksValidation. If you do not wish to perform validation (it is time consuming), set to False the variables performValidationOnSamplesThroughoutTraining
and performFullInferenceOnValidationImagesEveryFewEpochs.
h) (optional) If you need to adjust the length of the training session, eg for a smaller network, easiest way is to lower the total number of epochs numberOfEpochs=35. You should also then adjust the pre-defined schedule via predefinedSchedule. Another option is to use a decreasing schedule for the learning rate, by setting typeOfLearningRateSchedule = 'poly'.
i) To test a trained network, you need to point to the images of the testing subjects, similar to point d) for the training. Adjust the variable channels = ["./testChannels_flair.cfg", "./testChannels_t1c.cfg"] to point to the modalities of the test subjects. If ROI masks are available, point to them via roiMasks and inference will only be performed within the ROI. Else comment this variable out. Similarly, if you provide the ground-truth labels for the testing subjects via gtLabels, accuracy of the prediction will be calculated and the DSC metric will be reported. Otherwise just comment this variable out.
j) Finally, you need to create a file, which will list names to give to the predictions for each of the testing subject. See entry namesForPredictionsPerCase = "./testNamesOfPredictionsSimple.cfg" and the corresponding pre-set file. After that, you are ready to test with a model.
The provided configuration of the DeepMedic takes roughly 2 days to get trained on an NVIDIA GTX Titan X. Inference on a standard size brain scan should take 2-3 minutes. Adjust configuration of training and testing or consider downsampling your data if it takes much longer for your task.
We hope that making this software publically available can accelerate adoption of Deep Learning in the field of Biomedical Image Processing and Analysis. It is still actively developed, so do take into account that it is far from modular and fully generic. In its current state, we believe it can serve as a helpful baseline method for the benchmarking of further learners, as well as a segmentation system for various pipelines. If you find our work has positively influenced yours, please cite our paper [1].
I am well aware that much of the functionality is not fully modular and the API far from perfect. I hope to make DeepMedic a helpful tool for the community. So feel free to email me with your feedback or any issues at: konstantinos.kamnitsas12@ic.ac.uk
Best wishes,
Konstantinos Kamnitsas
License for the DeepMedic software: BSD 3-Clause License. A copy of this license is present in the root directory.
License for the data provided for the examples: Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Switzerland License. http://creativecommons.org/licenses/by-nc-sa/3.0/ch/deed.en