Tianshu Chu* (New York University), Xinmeng Li* (New York University), Huy V. Vo (Ecole Normale Superieure, INRIA and Valeo.ai), Ronald M. Summers (National Institutes of Health Clinical Center), Elena Sizikova (New York University) *- equal contribution.
This is a code release of the paper "Weakly Supervised Lesion Segmentation using Multi-Task Learning".
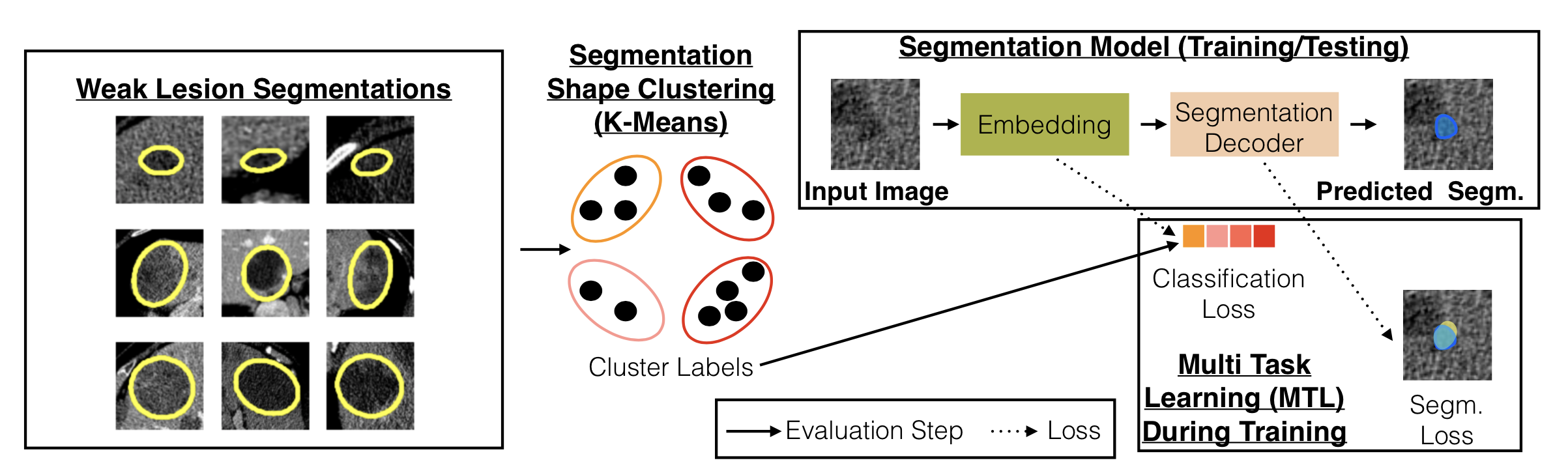
We introduce the concept of multi-task learning to weakly-supervised lesion segmentation, one of the most critical and challenging tasks in medical imaging. Due to the lesions' heterogeneous nature, it is difficult for machine learning models to capture the corresponding variability. We propose to jointly train a lesion segmentation model and a lesion classifier in a multi-task learning fashion, where the supervision of the latter is obtained by clustering the RECIST measurements of the lesions.
This work is released under the GNU General Public License (GNU GPL) license.
- python3.8 was tested
- pytorch 1.7.1 version was tested
- Please refer to the requirements.txt file for other dependencies.
- pip install -r requirements.txt
- git clone https://github.com/qubvel/segmentation_models.pytorch.git
- Raw images: https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/DBW86T
- Ground true segmentations: https://www.kaggle.com/tschandl/ham10000-lesion-segmentations?select=HAM10000_segmentations_lesion_tschandl
- For data preprocessing:
python code_ham/HAM_data.py
Under code_ham/ directory
- To train A1 model in the paper:
python HAM_A1_train.py
- To train A1+L model in the paper:
python HAM_A1class_train.py
- To train Acoseg model in the paper:
python HAM_Acoseg_train.py
Under code_ham/ directory
- To evaluate A1 model in the paper:
python HAM_A1_eval.py
- To evaluate A1+L model in the paper:
python HAM_A1class_eval.py
- To evaluate Acoseg model in the paper:
python HAM_Acoseg_eval.py
Put under the folder code_ham/checkpoints to evaluate:
- A1: https://drive.google.com/file/d/12gK0K_SZNleFAFVwPjzspHIS3rsWGXEb/view?usp=sharing
- A1+L: https://drive.google.com/file/d/1yFIfjXay9TRw_Wn_QLbfAdhMpFhcAqdC/view?usp=sharing
- ACoseg: https://drive.google.com/file/d/1B-3bG26yqpnupkH-q-qNHp1_IrU4EQf4/view?usp=sharing
- Unzip the raw images and segmentations to code_lits/data/volume/ and code_lits/data/segmentations/: https://www.kaggle.com/andrewmvd/liver-tumor-segmentation https://www.kaggle.com/andrewmvd/liver-tumor-segmentation-part-2?select=volume_pt6
- For data preprocessing:
python code_lits/lits_data.py
Under code_lits/ directory
- To train A1 model in the paper:
python lits_A1_train.py
- To train A1+L model in the paper:
python lits_A1class_train.py
- To train Acoseg model in the paper:
python lits_Acoseg_train.py
Under code_lits/ directory
- To evaluate A1 model in the paper:
python lits_A1_eval.py
- To evaluate A1+L model in the paper:
python lits_A1class_eval.py
- To evaluate Acoseg model in the paper:
python lits_Acoseg_eval.py
Put under the folder code_lits/checkpoints to evaluate:
- A1: https://drive.google.com/file/d/1EpQGDLOC8q95oqSvm0bZO6JdFSznCRKw/view?usp=sharing
- A1+L: https://drive.google.com/file/d/1OQFggEsLT2B84Na1iBITnmiip2dQULiw/view?usp=sharing
- ACoseg: https://drive.google.com/file/d/1vj4-vqEsfdET031Motq45I7KZRB6b8sC/view?usp=sharing
- For data preprocessing:
python code_deeplesion/DeepLesion_data.py
Under code_deeplesion/ directory
- To train A1 model in the paper:
python deeplesion_A1_train.py
- To train A1+L model in the paper:
python deeplesion_A1class_train.py
- To train Acoseg model in the paper:
python deeplesion_A1coseg_train.py
Under code_deeplesion/ directory
- To evaluate A1 model in the paper:
python deeplesion_A1_eval.py
- To evaluate A1+L model in the paper:
python deeplesion_A1class_eval.py
- To evaluate Acoseg model in the paper:
python deeplesion_A1coseg_eval.py
Put under the folder code_deeplesion/checkpoints to evaluate:
- A1: https://drive.google.com/file/d/1NlwB0dr_hCgpIdro6T4wrjAr1pO4ulFS/view?usp=sharing
- A1+L: https://drive.google.com/file/d/1V-NisPmQwvXy3TjawhFeEABIXtNxZfKQ/view?usp=sharing
- ACoseg: https://drive.google.com/file/d/1ygR4FZCwRkR0a87vzKeGqYEjl_n4A0tQ/view?usp=sharing
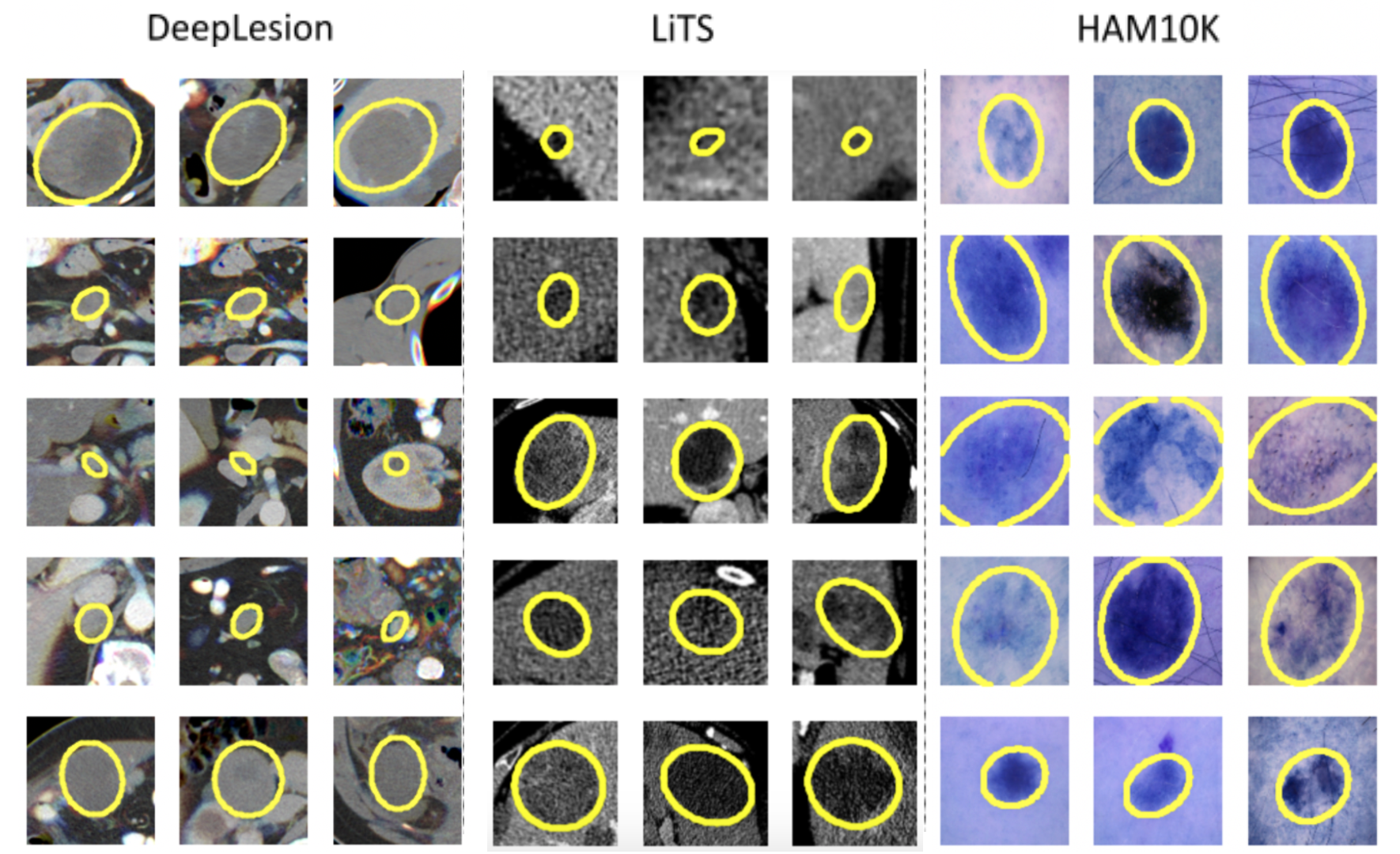
Cluster visualizations can be seen below. Clustering based on RECIST parameters allows grouping lesions into groups with similar shapes.
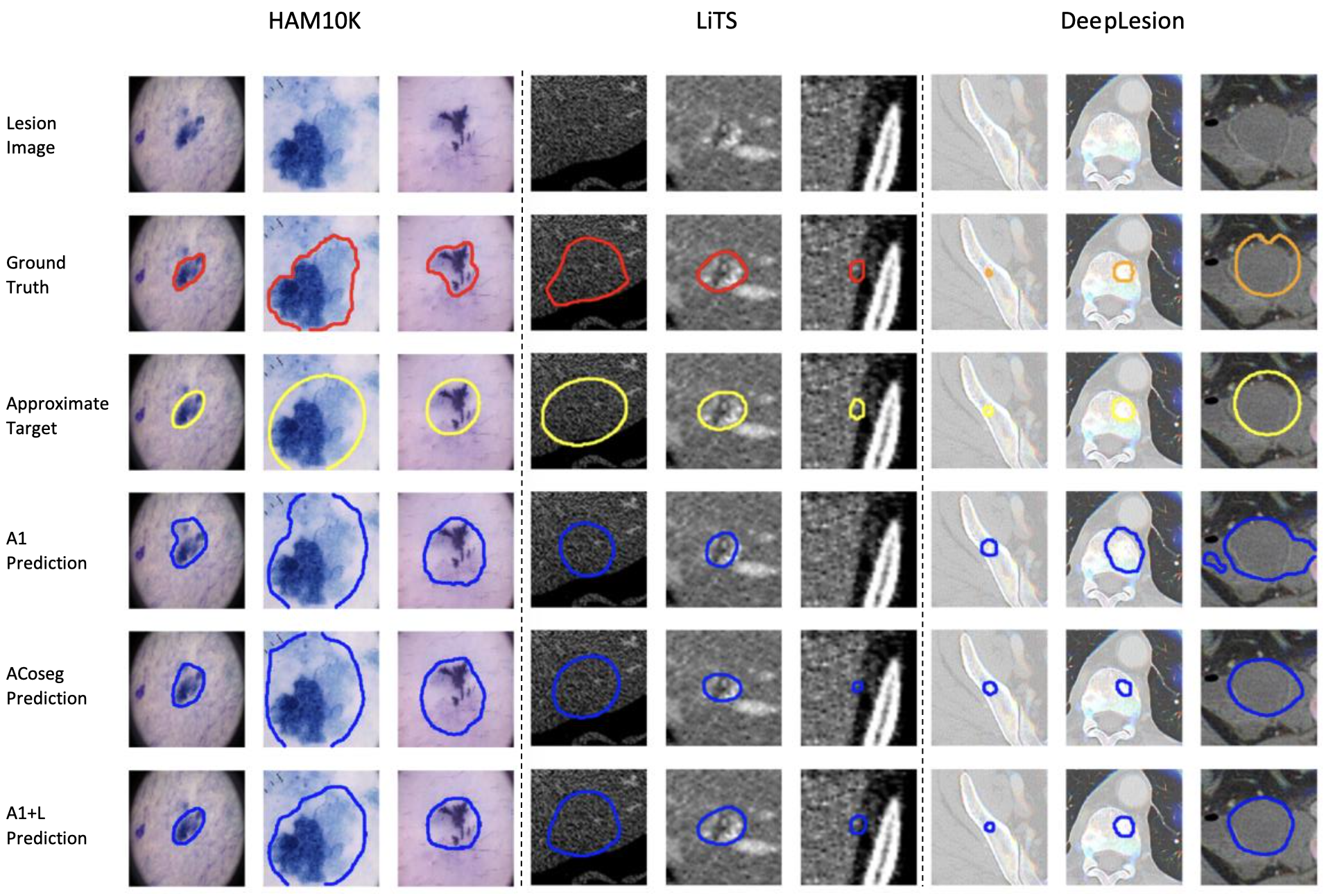
Visualization of sample segmentation results. 1st row - input lesion image. 2nd row - ground truth. 3rd row - appeoximate target. 4th row - result of A1 baseline, based on DeepLabV3+ (A1). 5th row - result of co-segmentation baseline (ACoseg), [Agarwal, 2020a]. 6th row - our result (A1+L).
@inproceedings{chuli2021lesion_wsol,
title={Improving Weakly Supervised Lesion Segmentation Using Multi-Task Learning},
author={Chu, Tianshu and Li, Xinmeng and Vo, Huy V. and Summers, Ronald M. and Sizikova, Elena},
booktitle={Proceedings of the Medical Imaging with Deep Learning (MIDL) Conference},
year={2021}
}