Enhanced Ligand Exploration and Interaction Recognition Algorithm (ELIXIR-A). ELIXIR-A, a plugin in VMD, can find similarities in the active site of the receptor under study compared to other receptors.
This is a Tcl/tk and python script for the identification of active sites and pharmacophores followed by ligand/inhibitor screening step. The package also provides an overall evaluation of the performance of the entire docking simulation.
The package has been tested on the following systems:
- MacOS: macOS Big Sur (recommended)
- Linux: Ubuntu 20.04 (recommended)
- Windows: 10 (partially compatible)
To run the script, you need to install python in your environment, python versions can be 3.7.4 or later.
The numpy package version 1.20.1 or later is required to run the algorithm.
The open3d package version 0.13.0 or later is required to run the algorithm.
The biopandas package version 0.28.0 or later is required to run the algorithm.
To install python on OS X & Linux:
sudo apt-get install python3To install numpy package on OS X & Linux:
sudo apt install python3-pip
pip install numpyIf python3 version is not compatible with Open3D package from PyPI, the . WHL package install file can be achieved from the Open3D official website.
For conda users, the latest version of Biopandas package can be achieved from the Biopandas official website.
pip install git+git://github.com/rasbt/biopandas.gitVMD 1.9.2 or later
In VMD, place the elixir2.1.1 folder to VMD TCL plugins directory /plugins/noarch/tcl/
Add the command in file /scripts/vmd/loadplugin.tcl.
Under line
### Modeling menu Add
vmd_install_extension elixir elixir_tk "Modeling/ELIXIR-A"If the package has been successfully installed, ELIXIR-A can be found under the Extensions-Modelling tab.
The total installation time will be about 30 minutes to install all the necessary packages.
The expected run time should be less than a minute.
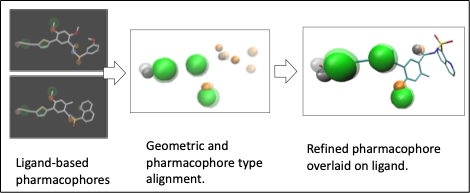
This demonstration uses ELIXIR-A to find refined pharmacophores from the RNA polymerase from dengue viruses 5I3P and 5I3Q.
Step 1. Load the first pharmacophore file 5I3P.pdb with Import pdb file icon at the first pharmacophore cluster area.
Step 2. Load the second pharmacophore file 5I3Q.pdb with Import pdb file icon at the second pharmacophore cluster area.
In VMD OpenGL Display window, the first pharmacophore cluster will be shown as Van der Waals balls and the second ones will be shown as red Van der Waals balls. The pharmacophore files can be generated by DruGUI plugin or interactive Pharmit platform using ligand/inhibitor binding structure.
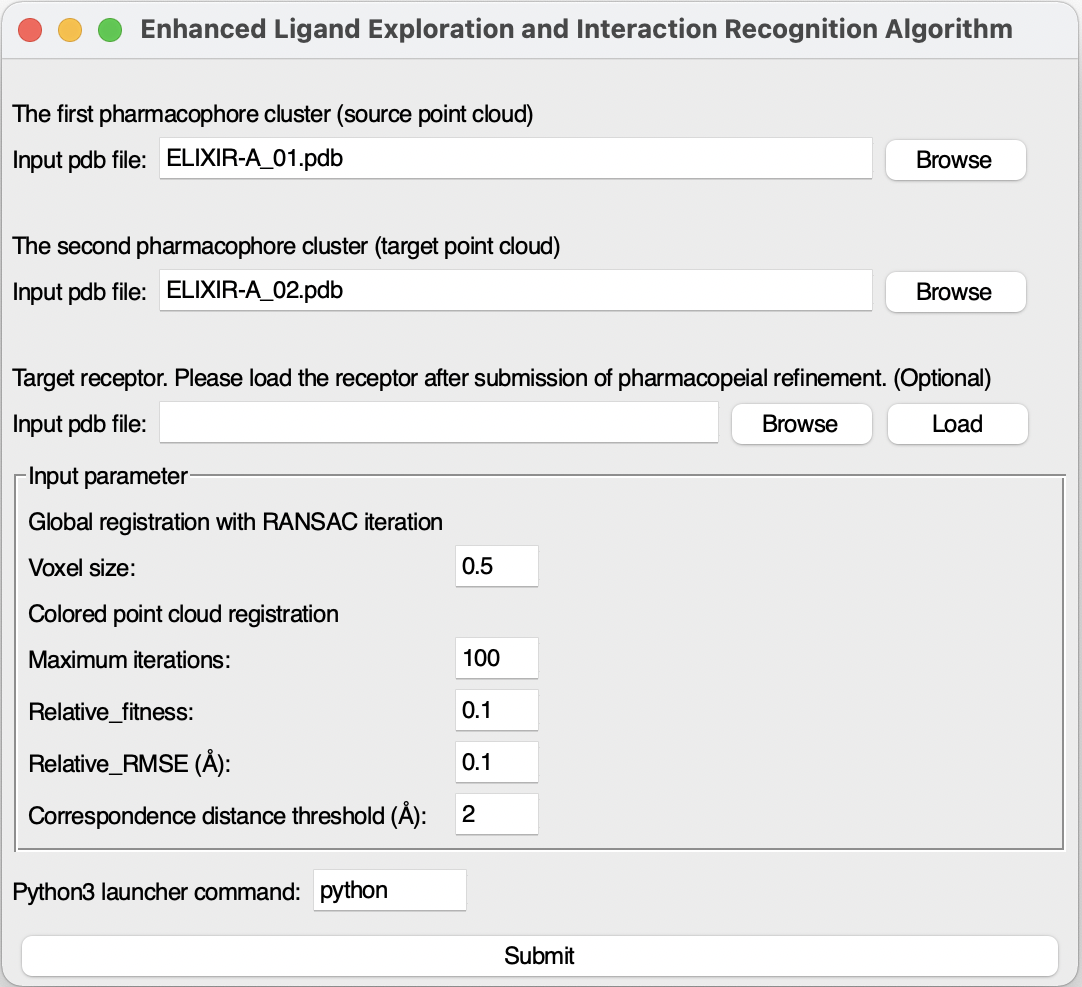
Pharmacophore clusters have been loaded.
Output pharmacophores have been generated and shown in VMD OpenGL Display window. It can also viewed by Schrödinger Maestro. Two pharmacophore clusters show some similar binding activities. And the refinement can be used for further drug screening.