Enhanced generalizability of RNA secondary structure prediction via convolutional block attention network and ensemble learning
The prediction of RNA secondary structure (RSS) is a fundamental but unmet need for RNA research. Various deep learning (DL)--based state-of-the-art (SOTA) methods achieved improved accuracy over thermodynamic-based methods. However, the over-parameterization nature of DL makes SOTA methods prone to overfitting and thus limits their generalizability. Meanwhile, the inconsistency of RSS predictions between SOTA methods further aggravated the crisis of generalizability.
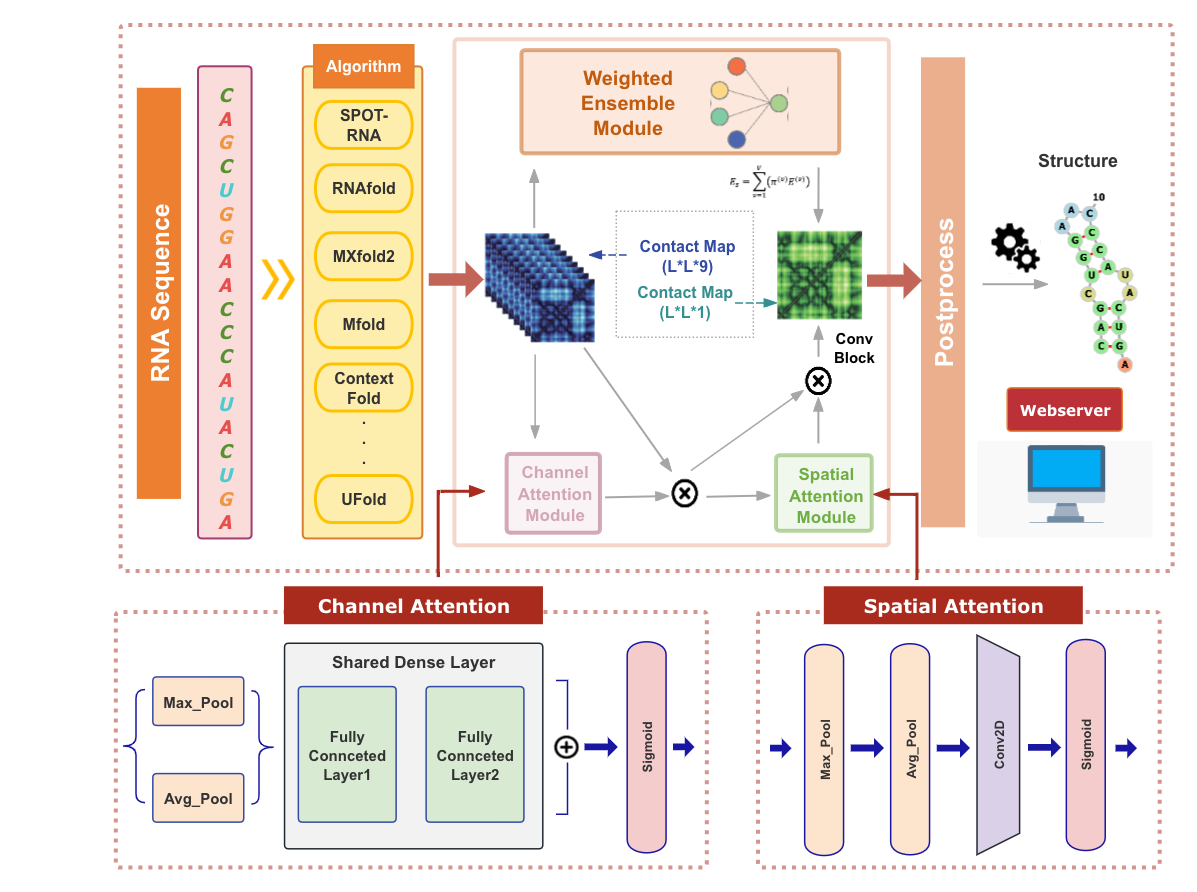
Here, we propose TrioFold to achieve enhanced generalizability of RSS prediction by integrating base-pairing clues learned from both thermodynamic- and DL-based methods by ensemble learning and convolutional block attention mechanism. TrioFold achieves higher accuracy in intra-family predictions and enhanced generalizability in inter-families and cross-RNA-types predictions. Importantly, TrioFold uses only ~2800 parameters to achieve superior performance over SOTA DL methods requiring millions of parameters. This study demonstrated new opportunities to enhance generalizability for RSS predictions by efficiently ensemble learning of base-pairing clues learned from both thermodynamic- and DL-based algorithms.
Before you begin, ensure you have met the following requirements:
Python >= 3.10
PyTorch >= 1.11
Subprocess
collections
numpy >= 1.23.5
- Clone the whole repository.
git clone https://github.com/sfsdfd62/TrioFold.git - Activate the conda environment.
conda activate TrioFold
We provide a script for testing and evaluating the prediction result. By running the following code, you will obtain results consistent with those reported in the paper. After installation, please download dataset files from Zenodo, put them into the /data/ folder, and change the file name in the python script.
test_data = RNASSDataGenerator('/dataset/','bpRNAnew.cPickle')
After the above procedure, just simply run
python TrioFold.py
You are expected to get the same results as we present in the paper.
However, due to copyright issues, we are unable to provide the installation program and steps for the base learners here. If you need prediction services, we strongly recommend using our webserver.
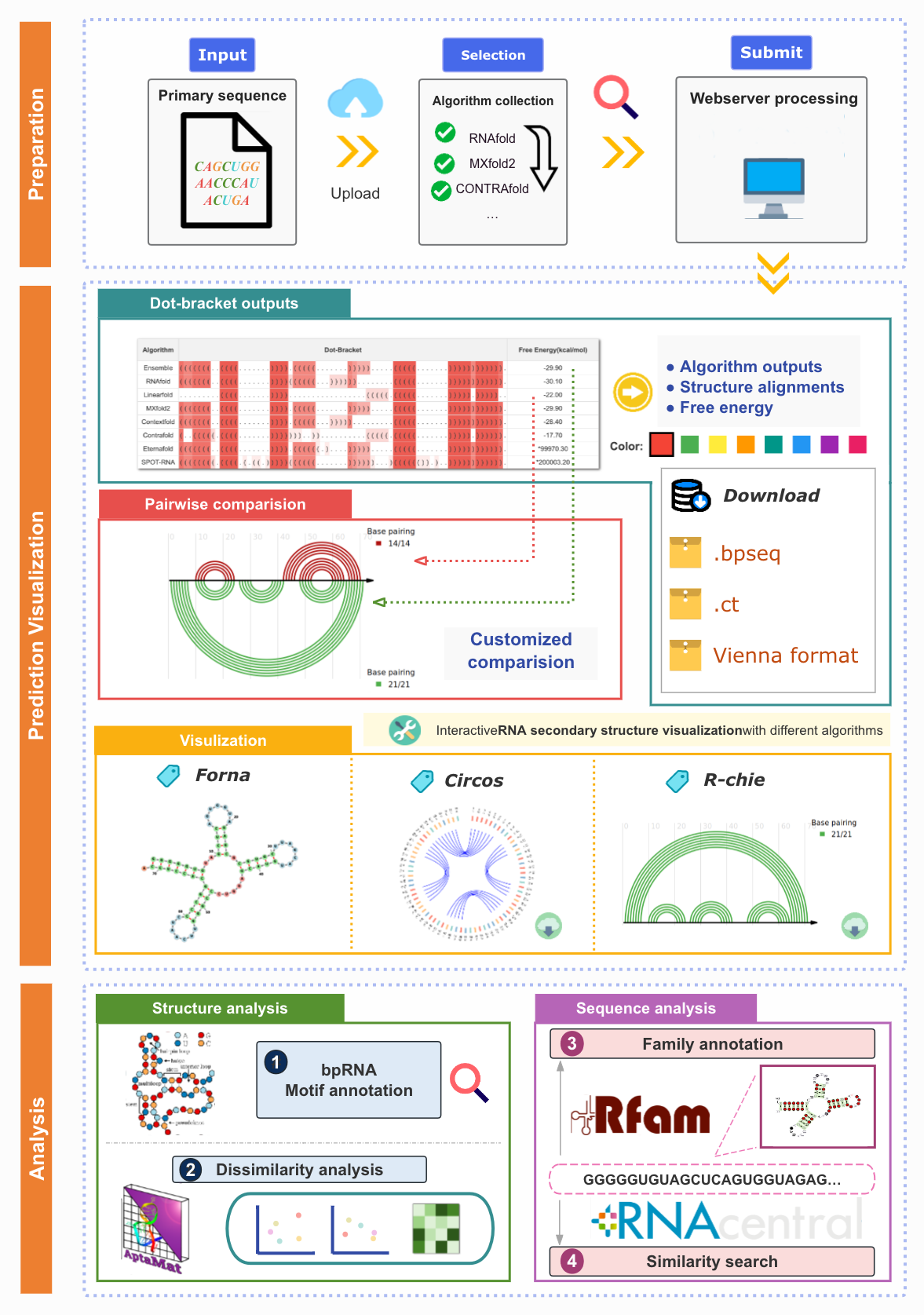
Moreover, we implemented TriFold and those base-learner methods in a one-stop user-friendly webserver to enable convenient usage for biologists without any programming requirement. The webserver provides RSS prediction and analysis functions and it can be freely accessed at http://triofold.aiddlab.com/.
Distributed under the MIT License. See LICENSE for more information.