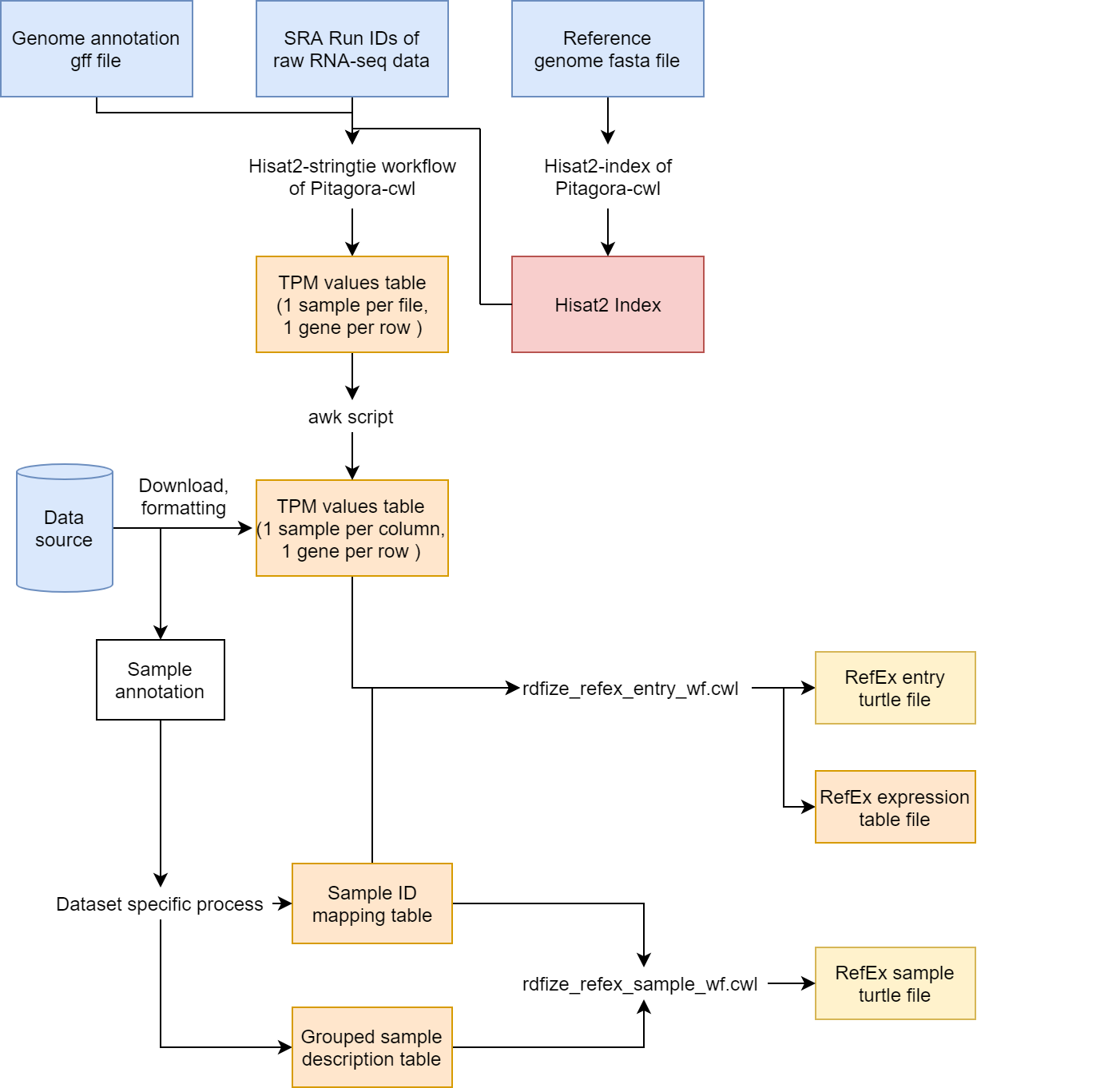
This document outlines the steps involved in preparing gene expression data presented on RefEx.
You can find the CWL workflows and tools used for the procedures in the cwl/ directory. The workflows output turtle format RDF files and tsv files.
Directories within the project/ directory contain documents to describe data sources for each project and the specific formatting processes for each dataset. These directories also include yml files for the CWL workflows.
If calculated TPM value data was provided by a project, we used the calculated data for RefEx.
If TPM value data was not available, we calculated the TPM values from the raw sequence data with HISAT2-StringTie workflow of Pitagora-cwl.
The workflow requires a list of SRA Run IDs as an input and calculates TPM values for every individual sample and gene.
The workflow also requires HISAT2 index files and a genome annotation file of the target species. The index files can be built from a genome sequence fasta file with the HISAT2-index tool of Pitagora-cwl.
The output files of the workflow are formatted as a TPM table, whose format is described in the next section.
-
TPM table
An input for
rdfize_refex_entry_wf.cwl. This is a tsv file containing TPM expression values.
Each row name is a gene ID. Any kind of gene ID (e.g. Ensembl, entrez, etc.) is allowed.
Each column name is a BioSample ID. If technical replicates exist and BioSample IDs are not unique within the dataset, SRA Run IDs may be used instead.
Table values are TPM of a sample specified by the column name and a gene specified by the row name.Example:
| SAMN07187967 | SAMN07187968 | SAMN07188034 | |
|---|---|---|---|
| FBgn0265945 | 0.0 | 0.0 | 0.126749 |
| FBgn0265946 | 0.0 | 0.0 | 4.181805 |
| FBgn0265947 | 0.0 | 0.0 | 0.0 |
| FBgn0003187 | 3.695364 | 0.628946 | 0.981611 |
-
Grouped sample description table
Input for
rdfize_refex_sample_wf.cwl.In RefEx, replicated samples are grouped and statistical values of TPM are calculated for each group.
This tsv table describes the annotation of the groups.
The columns 1 through 3 are:
RefexSampleId: ID for each sample group, which hasRESas the prefix.
Description: A human-readable description of a sample group.
NumberOfSamples: The number of samples which the sample group includes.
In columns 4 and after, any sample annotations can be described for each data set.
Example:
| RefexSampleId | Description | NumberOfSamples | Category | Strain | Tissue | Sex |
|---|---|---|---|---|---|---|
| RES00001615 | orgR, abdomen without digestive or reproductive system, female | 4 | tissues | orgR | abdomen without digestive or reproductive system | female |
| RES00001616 | orgR, abdomen without digestive or reproductive system, male | 4 | tissues | orgR | abdomen without digestive or reproductive system | male |
| RES00001617 | orgR, digestive plus excretory system, female | 4 | tissues | orgR | digestive plus excretory system | female |
| RES00001618 | orgR, digestive plus excretory system, male | 4 | tissues | orgR | digestive plus excretory system | male |
-
Sample ID mapping table
An input for both
rdfize_refex_entry_wf.cwlandrdfize_refex_sample_wf.cwl.
This table contains BioSample IDs, IDs given by each project, and RES IDs of the sample group to which the sample belong.
Statistics of TPM values are calculated for sample groups defined by this table.Column 1:
RefexSampleId
Column 2:BiosampleId
Column 3:ProjectSampleIdID given by each project.Example:
(In this example Project sample IDs are GEO sample IDs)
| RefexSampleId | BiosampleId | ProjectSampleId |
|---|---|---|
| RES00001615 | SAMN07187968 | GSM2647254 |
| RES00001615 | SAMN07187967 | GSM2647255 |
| RES00001615 | SAMN07188041 | GSM2647256 |
| RES00001616 | SAMN07188040 | GSM2647257 |
| RES00001616 | SAMN07188039 | GSM2647258 |
| RES00001616 | SAMN07188038 | GSM2647259 |
Besides a turtle file, rdfize_refex_entry_wf.cwl outputs a tsv table file which contains statistical values calculated for each sample group.
The statistical values are five-number summary (the minimum, the maximum, the median, and the first and third quartiles), the mean, and the standard deviation of TPM values and log2(TPM+1) values of samples in a sample group.
Note that the mean value of the log-transformed values is calculated after log-transformation of raw TPM values (i.e., it is not the log-transformed mean value).
rdfize_refex_sample_wf.yml.sample
eachsample_table_file:
class: File
path: /path/to/eachsample.tsv
sample_table_file:
class: File
path: /path/to/sample.tsv
id_uri_prefix: _URI_PREFIX_
id_uri_abbrev: _URI_ABBREV_
signature_file:
class: File
path: /path/to/signature.ttl
final_output_filename: refexsample_project_date.ttl
eachsample_table_file and sample_table_file are the path to the sample ID mapping table and the grouped sample description table, respectively.
id_uri_prefix is the prefix of project sample IDs' URI.
e.g. <http://fantom.gsc.riken.jp/5/sstar/FF:>
id_uri_abbrev is the abbreviation of the prefix indicated by id_uri_prefix.
e.g. ff
These will be output to the turtle as:
@prefix ff: <http://fantom.gsc.riken.jp/5/sstar/FF:> .
signature_file is the path to the turtle file which contains basic information of the RefEx RDF.
final_output_filename is the name of the output turtle file.
rdfize_refex_entry_wf.yml.sample
eachsample_table_file:
class: File
path: /path/to/eachsample.tsv
tpm_table_file:
class: File
path: /path/to/tpm.tsv
entry_id_num: _ID_TO_BEGIN_WITH_
entry_table_filename: project_refextable_entry.tsv
id_uri_prefix: _URI_PREFIX_
id_uri_abbrev: _URI_ABBREV_
signature_file:
class: File
path: /path/to/signature.ttl
concatenated_filename: refex_project_date.ttl
eachsample_table_file and tpm_table_file are the path to the sample ID mapping table and the tpm table, respectively.
entry_id_num is the number to be given to the first entry as its RFX ID.
id_uri_prefix is the prefix of gene IDs' URI.
e.g. <http://rdf.ebi.ac.uk/resource/ensembl/>
id_uri_abbrev is abbreviation of the prefix indicated by id_uri_prefix.
e.g. ensembl
These will be output in the turtle as:
@prefix ensembl: <http://rdf.ebi.ac.uk/resource/ensembl/> .
signature_file is the path to the turtle file which contains basic information of the RefEx RDF.
concatenated_filename is the name of the output turtle file.
$ cwltool rdfize_refex_entry_wf.cwl rdfize_refex_entry_wf.yml
$ cwltool rdfize_refex_sample_wf.cwl rdfize_refex_sample_wf.yml