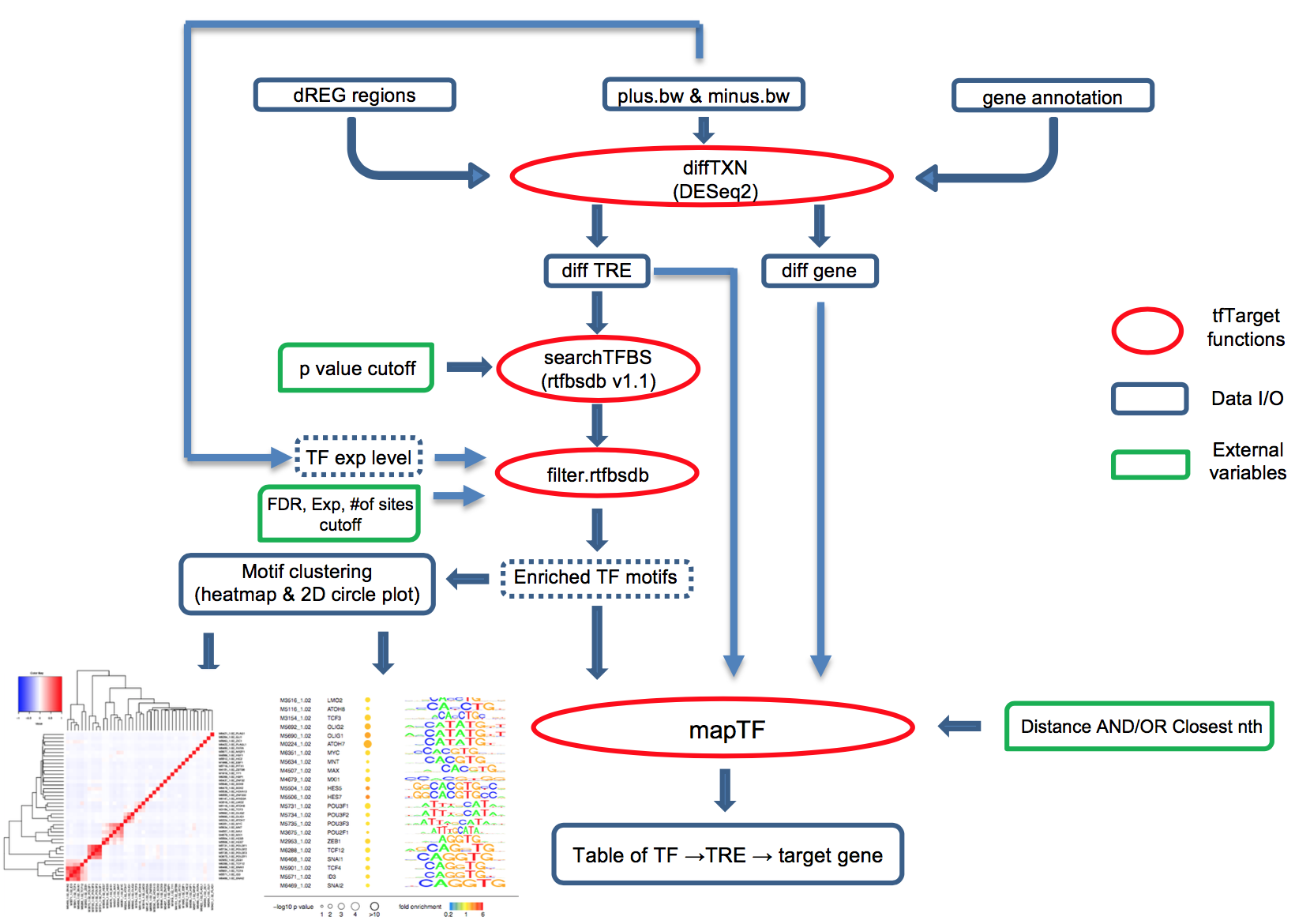
Transcription factors (TFs) regulate complex programs of gene transcription by binding to short DNA sequence motifs within transcription regulatory elements (TRE). Here we introduce tfTarget, a unified framework that identifies the "TF -> TRE -> target gene" networks that are differential regulated between two conditions, e.g. experimental vs. control, using PRO-seq/GRO-seq/ChRO-seq data as the input. The package provies a convenient interface for users without assuming knowledge with R environment, users can directly run the scipts in linux console.
Chromatin run-on reveals the transcriptional etiology of glioblastoma multiforme
Tinyi Chu, Edward J Rice, Gregory T Booth, Hans H Salamanca, Zhong Wang, Leighton J Core, Sharon L Longo, Robert J Corona, Lawrence S Chin, John T Lis, Hojoong Kwak, Charles Danko
bioRxiv 185991; doi: https://doi.org/10.1101/185991
-
R packages:
rphast, rtfbdbs, grid, cluster, apcluster, DESeq2, gplots.
rtfbsdb (https://github.com/Danko-Lab/rtfbs_db)
-
bioinformatics tools or exterior command:
awk, sort: Unix commands
bedtools (http://bedtools.readthedocs.org/en/latest/)
sort-bed (http://bedops.readthedocs.org/en/latest/index.html)
twoBitToFa, faToTwoBit (http://hgdownload.cse.ucsc.edu/admin/exe/)
-
2bit files for your genome of interest. Find links to these here:
-
tfs object file for the species of interests, in .rdata format, which contains the curated transcription factor motifs database. For Homo_sapiens, it is provided by tfTarget package, and will be used by default. For others species, we provide a convenient script get.tfs.R to call rtfbsdb, and generate the species.tfs.rdata.
example:
R --vanilla --slave --args Drosophila_melanogaster < get.tfs.RThe look-up table for species name can be found here: The "species" column (1st column) of http://cisbp.ccbr.utoronto.ca/summary.php?by=1&orderby=Species
-
TREs regions identified by dREG, or equivalent tools, in bed format.
To prepare the input TRE files, users are recommended to merge dREG sites from query and control samples, using bedtools merge (http://bedtools.readthedocs.io/en/latest/content/tools/merge.html), e.g.,
cat query.dREG.peak.score.bed control.dREG.peak.score.bed \
| LC_COLLATE=C sort -k1,1 -k2,2n \
| bedtools merge -i stdin > merged.dREG.bed
*Use zcat for bed.gz files.
-
Gene annotation file in bed6 format. Can be prepared from gencode or Refseq gtf files. We recommend to use gene ID and gene name for the 4th and 5th columns. The information will show up in the output. https://www.gencodegenes.org/releases/current.html
gtf.gz files can be converted to the gene annotation file for tfTarget input using the following command as an example:
zcat gencode.v19.annotation.gtf.gz \
| awk 'OFS="\t" {if ($3=="gene") {print $1,$4-1,$5,$10,$18,$7}}' \
| tr -d '";' > gencode.v19.annotation.bed
- bigWigs files of query and control replicates. The same requirement for preparing the input files for dREG. See this https://github.com/Danko-Lab/dREG#data-preparation
- If all dependent packages and commands have been installed, please use the following codes to install/update the package.
library("devtools");
install_github("Danko-Lab/tfTarget/tfTarget")
To use the tfTarget package, after installing tfTarget package, simply run "R --vanilla --slave --args ... < main.R" under the directory of main.R, with ... specifying arguments for tfTarget detailed as below.
Required arguments:
-query: query file names of PRO-seq in bigWig format, ordered by plus and minus pairings,
e.g. query1.plus.bw query1.minus.bw query2.plus.bw query2.minus.bw ...
(The default directory is the current working directory, use -bigWig.path to speficy if otherwise.)
-control: control file names of PRO-seq in bigWig format, ordered by plus and minus pairings,
e.g. control1.plus.bw control1.minus.bw control2.plus.bw control2.minus.bw ...
(The default directory is the current working directory, use -bigWig.path to speficy if otherwise.)
-prefix: prefix for the output pdfs and txts.
-TRE.path: input TRE regions, e.g. dREG sites, in bed3 format. Only the first three columns will be used.
-gene.path: Gene annotation file in bed6 format. Can be prepared from gencode or Refseq gtf files.
We recommend to use gene ID and gene name for the 4th and 5th columns.
The information will show up in the output. https://www.gencodegenes.org/releases/current.html
-2bit.path: 2bit files for your genome of interest.
Find links to these here: http://hgdownload.cse.ucsc.edu/downloads.html
Optional arguments:
Optional system arguments:
-bigWig.path: path to the bigWig files.
default="./"
-ncores: number of threads to use.
default=1.
-deseq: Use this tag indicates to run DEseq2 only.
No arugment is required. default is off.
-rtfbsdb: Use this tag indicates to run DEseq2 and then rtfbsdb only.
No arugment is required. default is off.
Optional DEseq2 arguments:
-pval.up: adjusted pvalue cutoff below which indicates differentially transcribed TREs.
default=0.01.
-pval.down: adjusted pvalue cutoff above which indicates TREs that are not significantly changed between query and control.
default=0.1
Optional rtfbsdb arguments:
-tfs.path: use this tag to specify tfs object from non-Homo sapiens species.
Can be prepared using get.tfs.R. See the "requires" section above.
-cycles: how many cycles to run GC-subsampled motif enrichment test.
default=2.
-mTH: threshold over which the TF motif is defined as significant different from the HMM background.
default=7.
-fdr.cutoff: cutoff of the median of pvalues from multiple GC-subsampled runs, above which defines significantly enriched motifs.
Optional mapTF arguments:
-dist: the distance cutoff for asscoiating TRE to the nearest annotated transcriptional start site.
default=1E6.
-closest.N: use this tag to report only the first nth genes to the TRE, can be used in combination with -dist.
default is off.
The output of an complete run of the main.R function will output .pdf files and .txt files.
Specifically,
1) .cor.heatmap.pdfs for TF motifs clustered by genomic locations,
2) .motif.ordered.pdfs for the visualization of TF motifs and their enrichment statistics
ordered by clusters in 1).
3) .TRE.deseq.txt for each TREs and their DESeq2 statistics.
4) .gene.deseq.txt for each annotated gene body and their DESeq2 statistics.
Rows with all NA value means that the length of gene is too short (<=1Kb) to be included for DESeq2 runs.
5) .TF.TRE.gene.txt for each TF whose motif is enriched in up/down TREs,
and the TREs that contains the motif, and the putative target genes for the TRE.
We recommend users to further filter the target genes of log2foldchange (and the pvalues)
with the same diretion of change as the TRE by which it is regulated.
-
R vignette: (Coming soon)
-
R manual: (Coming soon)