This is the official implementation for the paper:
Pre-training via Denoising for Molecular Property Prediction (Spotlight @ ICLR 2023)
by Sheheryar Zaidi*, Michael Schaarschmidt*, James Martens, Hyunjik Kim, Yee Whye Teh, Alvaro Sanchez Gonzalez, Peter Battaglia, Razvan Pascanu, Jonathan Godwin.
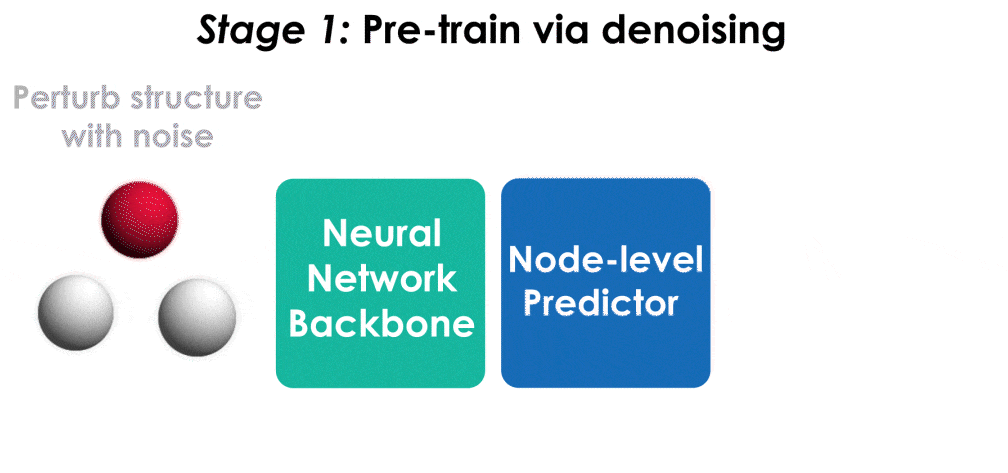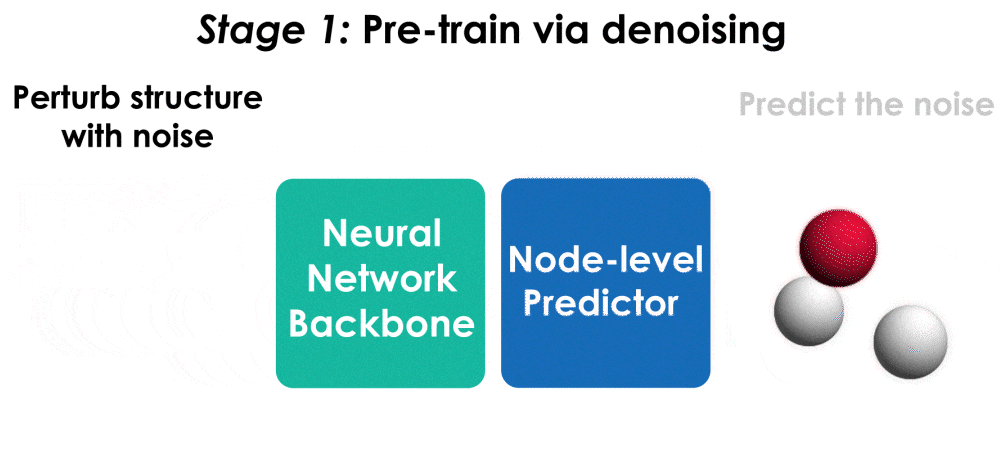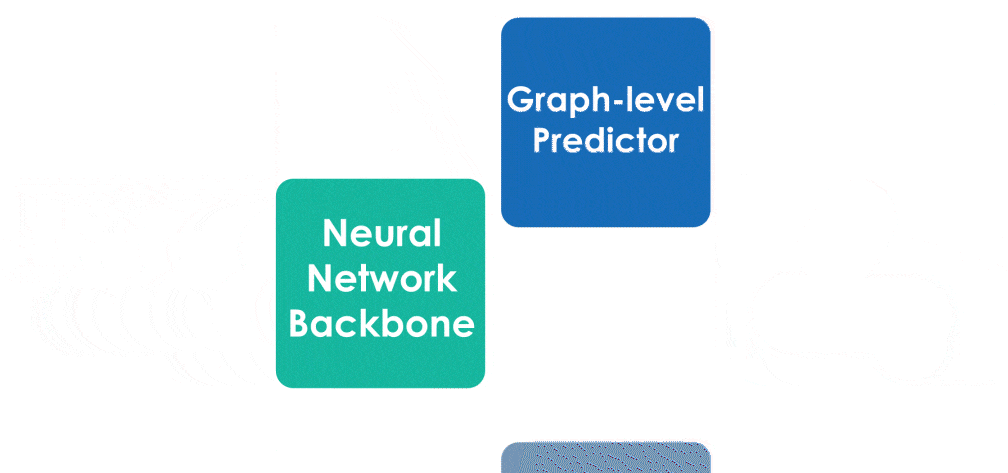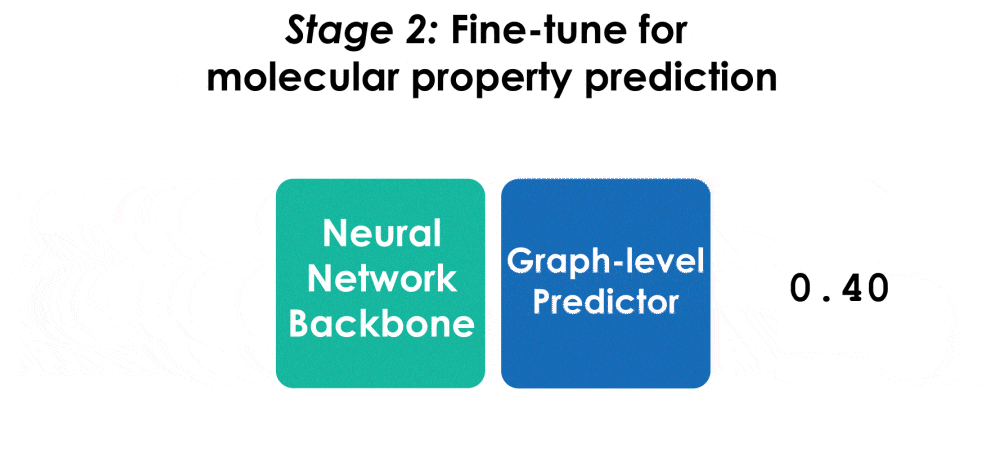
Pre-training via denoising is a powerful representation learning technique for molecules. This repository contains an implementation of pre-training for the TorchMD-NET architecture, built off the original TorchMD-NET repository.
Clone the repository:
git clone https://github.com/shehzaidi/pre-training-via-denoising.git
cd pre-training-via-denoising
Create a virtual environment containing the dependencies and activate it:
conda env create -f environment.yml
conda activate pvd
Install the package into the environment:
pip install -e .
The model is pre-trained on the PCQM4Mv2 dataset, which contains over 3 million molecular structures at equilibrium. Run the following command to pre-train the architecture first. Note that this will download and pre-process the PCQM4Mv2 dataset when run for the first time, which can take a couple of hours depending on the machine.
python scripts/train.py --conf examples/ET-PCQM4MV2.yaml --layernorm-on-vec whitened --job-id pretraining
The option --layernorm-on-vec whitened includes an optional equivariant whitening-based layer norm, which stabilizes denoising. The pre-trained model checkpoint will be in ./experiments/pretraining. A pre-trained checkpoint is included in this repo at checkpoints/denoised-pcqm4mv2.ckpt.
To fine-tune the model for HOMO/LUMO prediction on QM9, run the following command, specifying homo/lumo and the path to the pre-trained checkpoint:
python scripts/train.py --conf examples/ET-QM9-FT.yaml --layernorm-on-vec whitened --job-id finetuning --dataset-arg <homo/lumo> --pretrained-model <path to checkpoint>The fine-tuned model achieves state-of-the-art results for HOMO/LUMO on QM9:
| Target | Test MAE (meV) |
|---|---|
| HOMO | 15.5 |
| LUMO | 13.2 |
By default, the code will use all available GPUs to train the model. We used three GPUs for pre-training and two GPUs for fine-tuning (NVIDIA RTX 2080Ti), which can be set by prefixing the commands above with e.g. CUDA_VISIBLE_DEVICES=0,1,2 to use three GPUs.
It is straightforward to implement denoising in an existing codebase. There are broadly three steps:
- Add noise to the input molecular structures in the dataset. See here.
- Add an output module to the architecture for predicting the noise. See here.
- Use (or augment an existing loss with) an L2 loss for training the model. See here.
If you have found this work useful, please consider using the following citation:
@inproceedings{
zaidi2023pretraining,
title={Pre-training via Denoising for Molecular Property Prediction},
author={Sheheryar Zaidi and Michael Schaarschmidt and James Martens and Hyunjik Kim and Yee Whye Teh and Alvaro Sanchez-Gonzalez and Peter Battaglia and Razvan Pascanu and Jonathan Godwin},
booktitle={International Conference on Learning Representations},
year={2023},
url={https://openreview.net/forum?id=tYIMtogyee}
}