The repo is code (baseline and the proposed MetaPred) for paper MetaPred: Meta-Learning for Clinical Risk Prediction with Limited Patient Electronic Health Records by Xi Sheryl Zhang, Fengyi Tang, Hiroko H. Dodge, Jiayu Zhou, and Fei Wang.
MetaPred is a meta-learning framework for Clinical Risk Prediction using limited patient Electronic Health Records (EHRs). We given an example in the following figure:
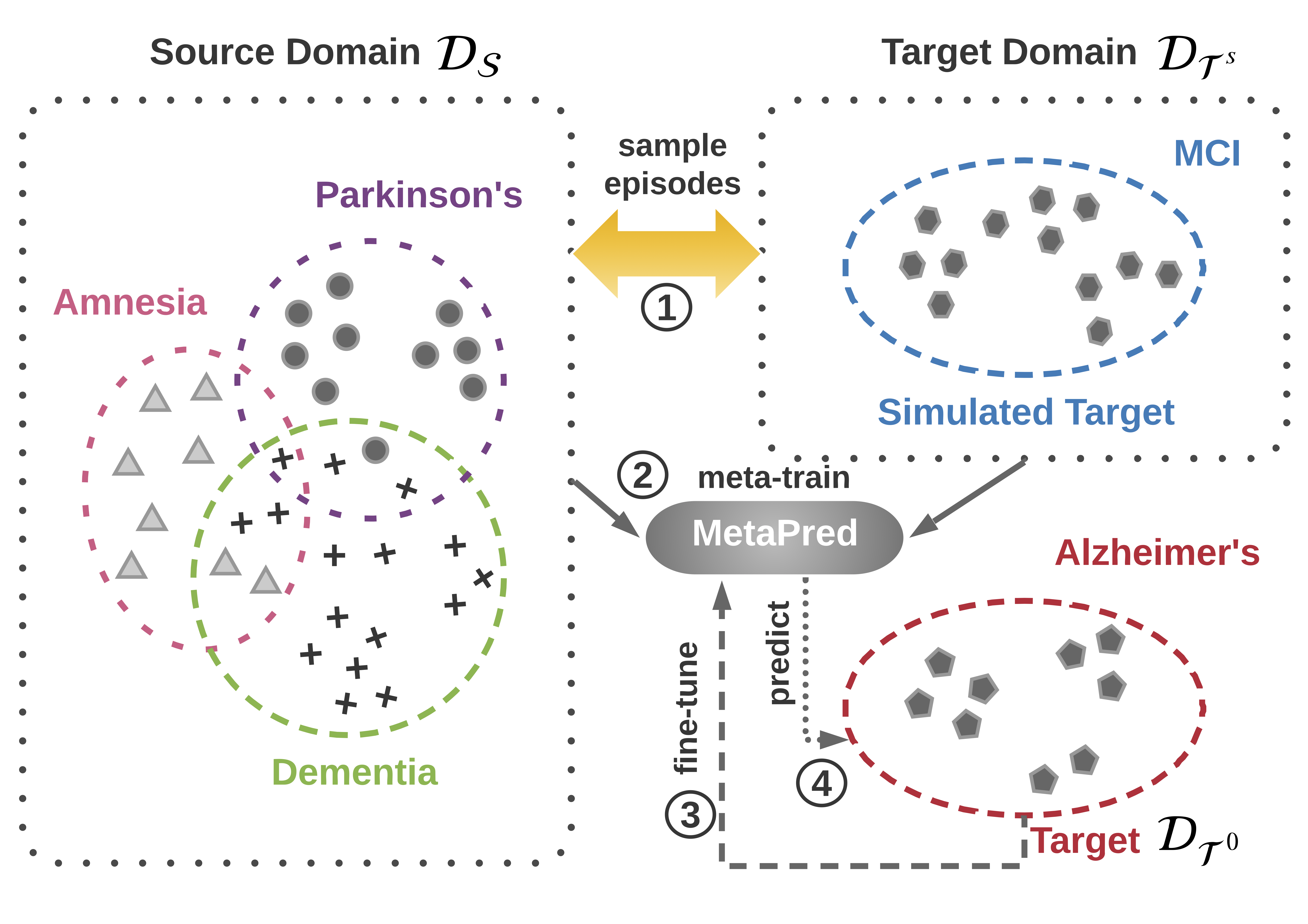
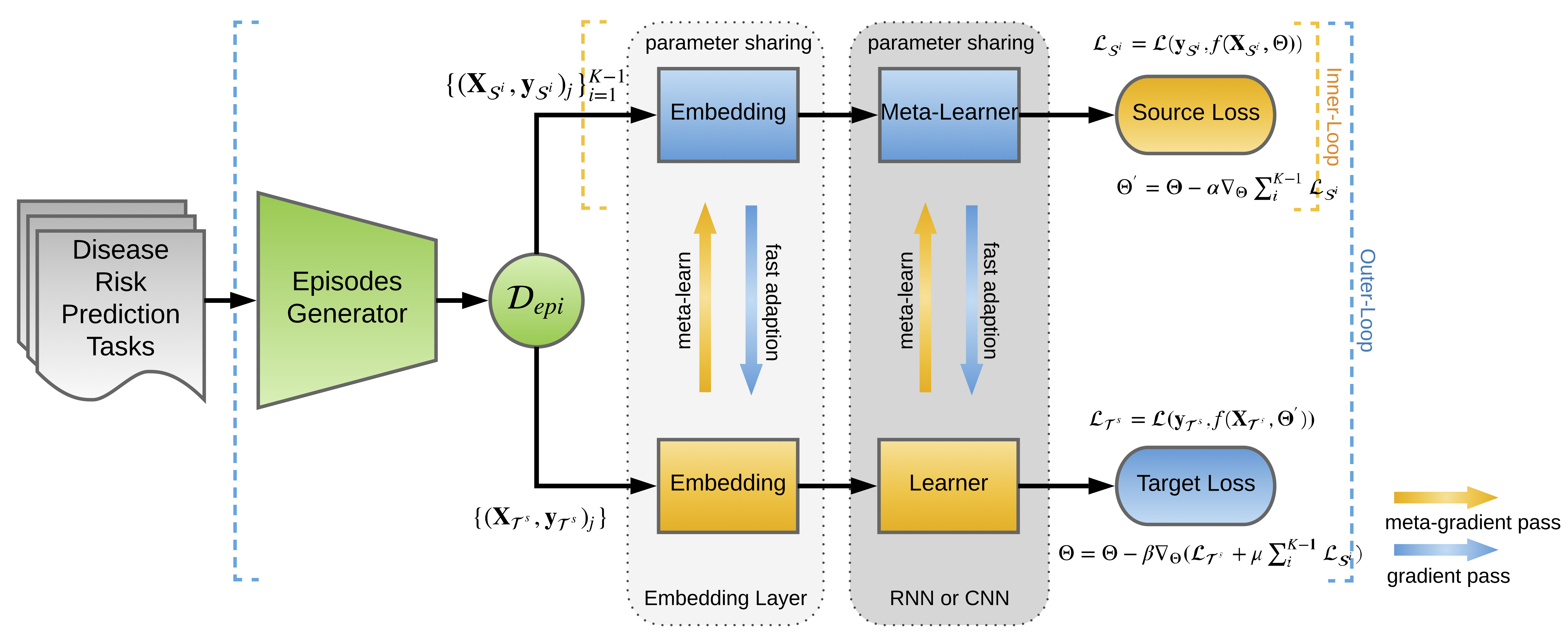
Suppose we have multiple domains, our goal is to predict Alzheimer’s disease with few labeled patients, which give rise to a low-resource classification. The idea is to employ labeled patients from high-resource domains and design a learning to transfer framework with sources and a simulated target in meta-learning. There are four steps: (1) constructing episodes by sampling from the source domains and the simulated target domain; (2) learn the parameters of predictors in an episode-by-episode manner; (3) fine-tuning the model parameters on the genuine target domain; (4) predicting the target clinical risk. We respectively implemented Convolutional Neural Network (CNN) and Long-Shot Term Memory (LSTM) Network as base predictors. The model overview (meta-training procedure) is shown as follows:
The entire learning procedure can be viewed as: iteratively transfer the parameter Θ learned from source domains through utilizing it as the initialization of the parameter that needs to be updated in the target domain.
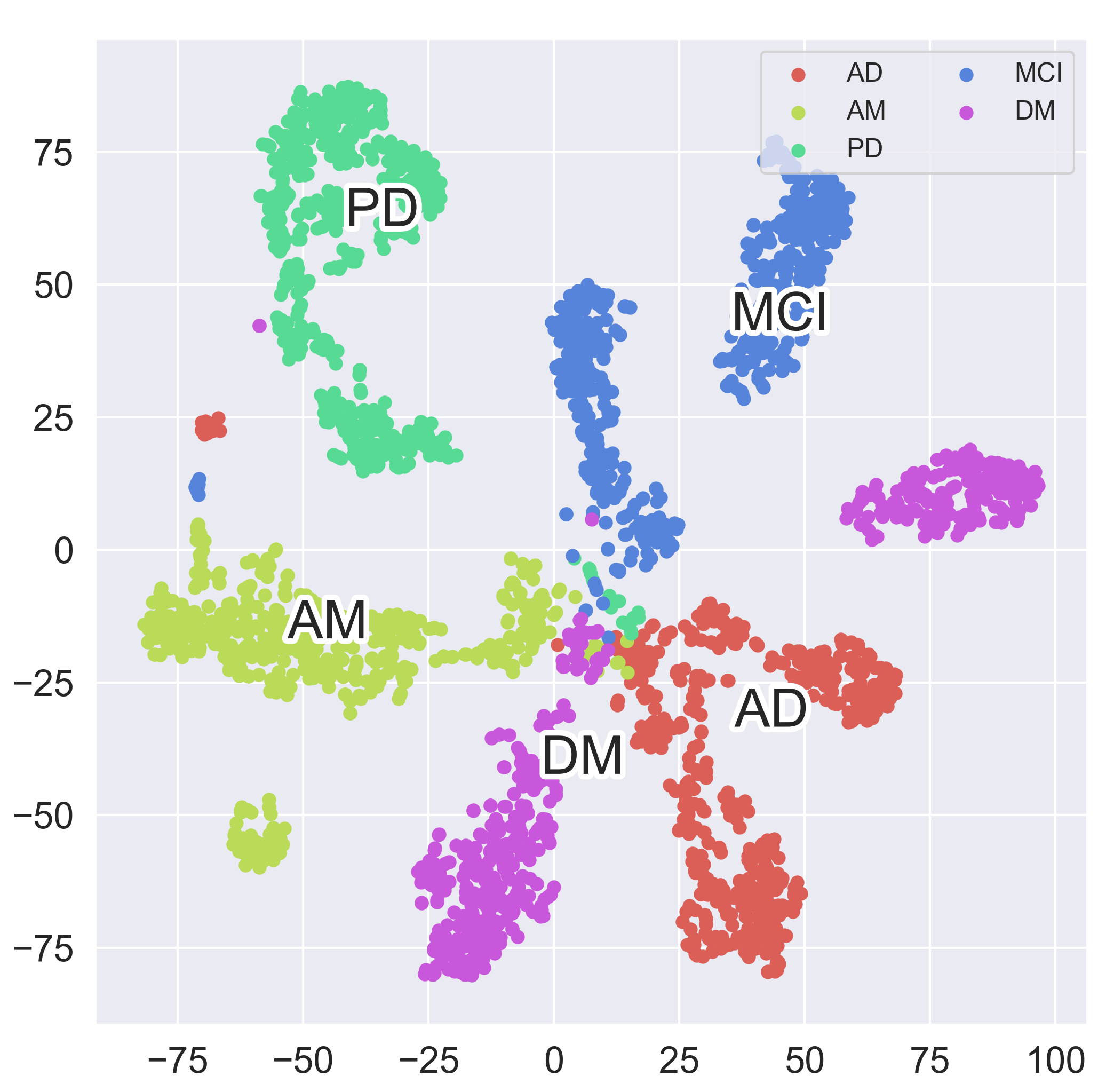
The learned representations of patients in five disease domains are visualized by t-SNE. In detail, AD, PD, DM, AM, MCI are abbreviations of Alzheimer's Disease, Parkinson's Disease, Dementia, Amnesia and Mild Cognitive Impairment, respectively. As a patient might suffer multiple diseases, there is supposed to be some overlaps among the given domains.
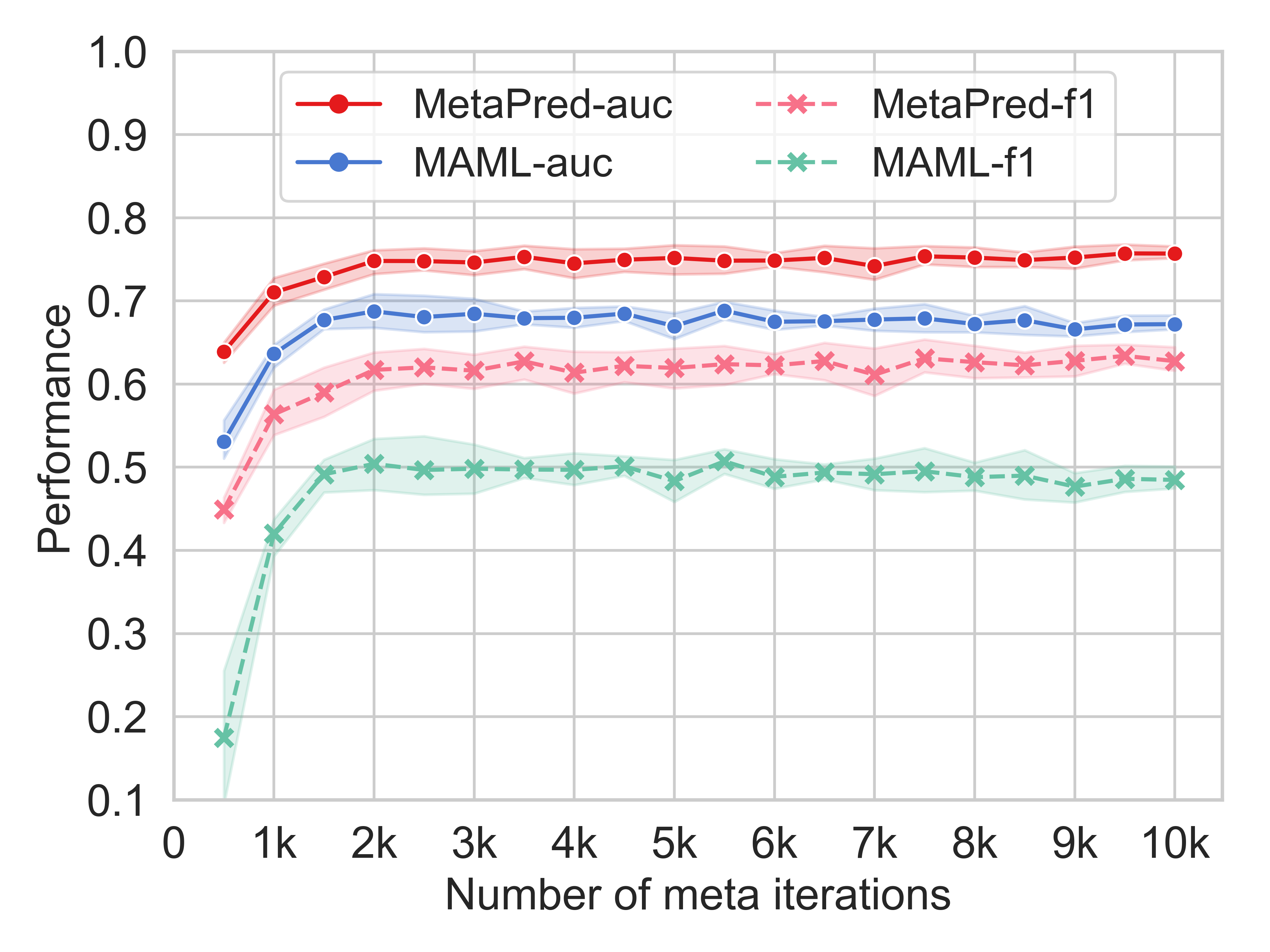
To demonstrate the effectiveness of the proposed MetaPred in the context of domain adaptation, we compare it with the state-of-the-art meta-learning algorithm Model-Agnostic Meta-Learning for Fast Adaptation of Deep Networks (MAML). The results on Alzheimer's Disease domain are presented in terms of AUC and F1-Score.
This package has the following requirements:
Python 3.x- TensorFlow 1.5
- Progress Bar
The baseline implementation includes:
- Logistic Regression
- K-Nearest Neighbors
- XGBoost
- SVM
- Random Forest
- MLP
- LSTM
- CNN
which can be used in any sort of sequence modeling, especially for EHRs data, directly.
To run MetaPred on EHR data, you need to revise the learning settings in main.py and the network hyperparameters in model.py. Then run the shell script metapred.sh.
bash metapred.shOur settings of learning procedures are:
python main.py --method='cnn' --metatrain_iterations=10000 --meta_batch_size=32 --update_batch_size=4 --meta_lr=0.001 --update_lr=1e-5 --num_updates=4 --n_total_batches=500000or
python main.py --method='rnn' --metatrain_iterations=10000 --meta_batch_size=32 --update_batch_size=4 --meta_lr=0.001 --update_lr=1e-5 --num_updates=4 --n_total_batches=500000There is implementations used in:
Chelsea Finn, Pieter Abbeel, Sergey Levine, Model-Agnostic Meta-Learning for Fast Adaptation of Deep Networks, International Conference on Machine Learning (ICML), 2017.
If you happen to use our work, please consider citing our paper:
@inproceedings{Zhang:2019:MMC:3292500.3330779,
author = {Zhang, Xi Sheryl and Tang, Fengyi and Dodge, Hiroko H. and Zhou, Jiayu and Wang, Fei},
title = {MetaPred: Meta-Learning for Clinical Risk Prediction with Limited Patient Electronic Health Records},
booktitle = {Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery \& Data Mining},
series = {KDD '19},
year = {2019},
location = {Anchorage, AK, USA},
pages = {2487--2495},
}
This paper can be accessed on : [MetaPred] (https://dl.acm.org/citation.cfm?id=3330779)