Implementation of
- MemStream: Memory-Based Anomaly Detection in Multi-Aspect Streams with Concept Drift . Siddharth Bhatia, Arjit Jain, Shivin Srivastava, Kenji Kawaguchi, Bryan Hooi
MemStream detects anomalies from a multi-aspect data stream. We output an anomaly score for each record. MemStream is a memory augmented feature extractor, allows for quick retraining, gives a theoretical bound on the memory size for effective drift handling, is robust to memory poisoning, and outperforms 11 state-of-the-art streaming anomaly detection baselines.
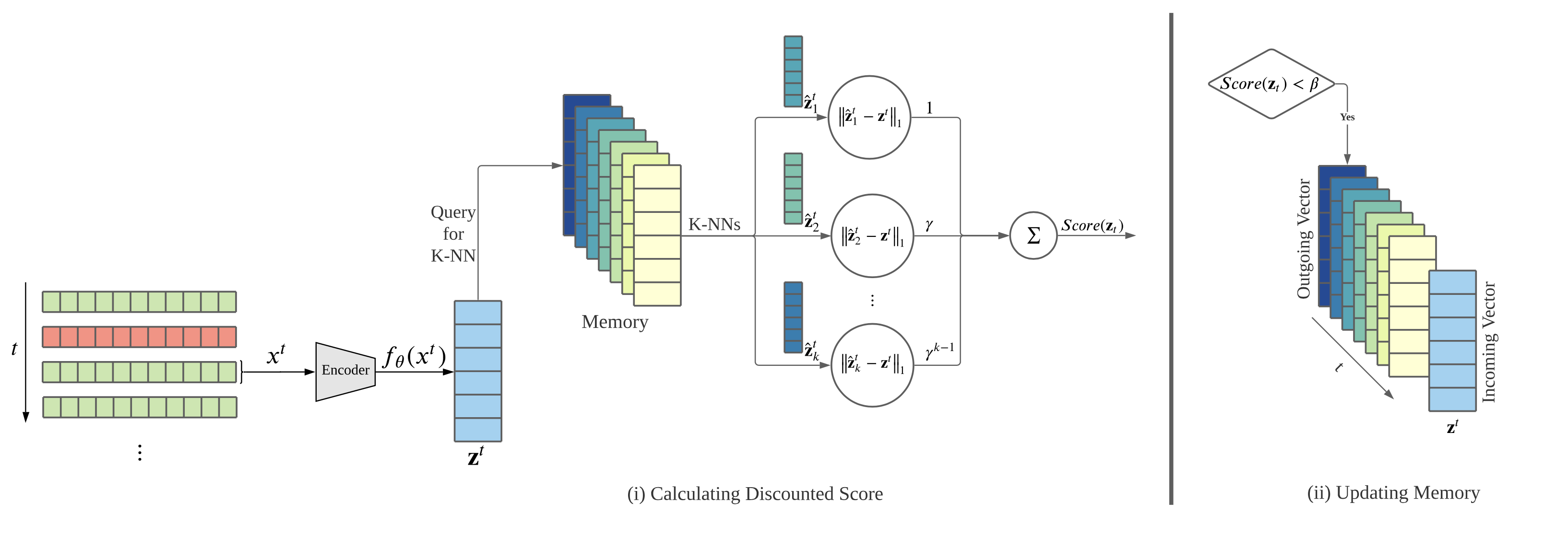 After an initial training of the feature extractor on a small subset of normal data, MemStream processes records in two steps: (i) It outputs anomaly scores for each record by querying the memory for K-nearest neighbours to the record encoding and calculating a discounted distance and (ii) It updates the memory, in a FIFO manner, if the anomaly score is within an update threshold β.
After an initial training of the feature extractor on a small subset of normal data, MemStream processes records in two steps: (i) It outputs anomaly scores for each record by querying the memory for K-nearest neighbours to the record encoding and calculating a discounted distance and (ii) It updates the memory, in a FIFO manner, if the anomaly score is within an update threshold β.
Processed Datasets can be downloaded from here. Please unzip and place the files in the data folder of the repository.
- KDDCUP99: Run
python3 memstream.py --dataset KDD --beta 1 --memlen 256 - NSL-KDD: Run
python3 memstream.py --dataset NSL --beta 0.1 --memlen 2048 - UNSW-NB 15: Run
python3 memstream.py --dataset UNSW --beta 0.1 --memlen 2048 - CICIDS-DoS: Run
python3 memstream.py --dataset DOS --beta 0.1 --memlen 2048 - SYN: Run
python3 memstream-syn.py --dataset SYN --beta 1 --memlen 16 - Ionosphere: Run
python3 memstream.py --dataset ionosphere --beta 0.001 --memlen 4 - Cardiotocography: Run
python3 memstream.py --dataset cardio --beta 1 --memlen 64 - Statlog Landsat Satellite: Run
python3 memstream.py --dataset statlog --beta 0.01 --memlen 32 - Satimage-2: Run
python3 memstream.py --dataset satimage-2 --beta 10 --memlen 256 - Mammography: Run
python3 memstream.py --dataset mammography --beta 0.1 --memlen 128 - Pima Indians Diabetes: Run
python3 memstream.py --dataset pima --beta 0.001 --memlen 64 - Covertype: Run
python3 memstream.py --dataset cover --beta 0.0001 --memlen 2048
--dataset: The dataset to be used for training. Choices 'NSL', 'KDD', 'UNSW', 'DOS'. (default 'NSL')--beta: The threshold beta to be used. (default: 0.1)--memlen: The size of the Memory Module (default: 2048)--dev: Pytorch device to be used for training like "cpu", "cuda:0" etc. (default: 'cuda:0')--lr: Learning rate (default: 0.01)--epochs: Number of epochs (default: 5000)
MemStream expects the input multi-aspect record stream to be stored in a contains , separated file.
- KDDCUP99
- NSL-KDD
- UNSW-NB 15
- CICIDS-DoS
- Synthetic Dataset (Introduced in paper)
- Ionosphere
- Cardiotocography
- Statlog Landsat Satellite
- Satimage-2
- Mammography
- Pima Indians Diabetes
- Covertype
This code has been tested on Debian GNU/Linux 9 with a 12GB Nvidia GeForce RTX 2080 Ti GPU, CUDA Version 10.2 and PyTorch 1.5.

