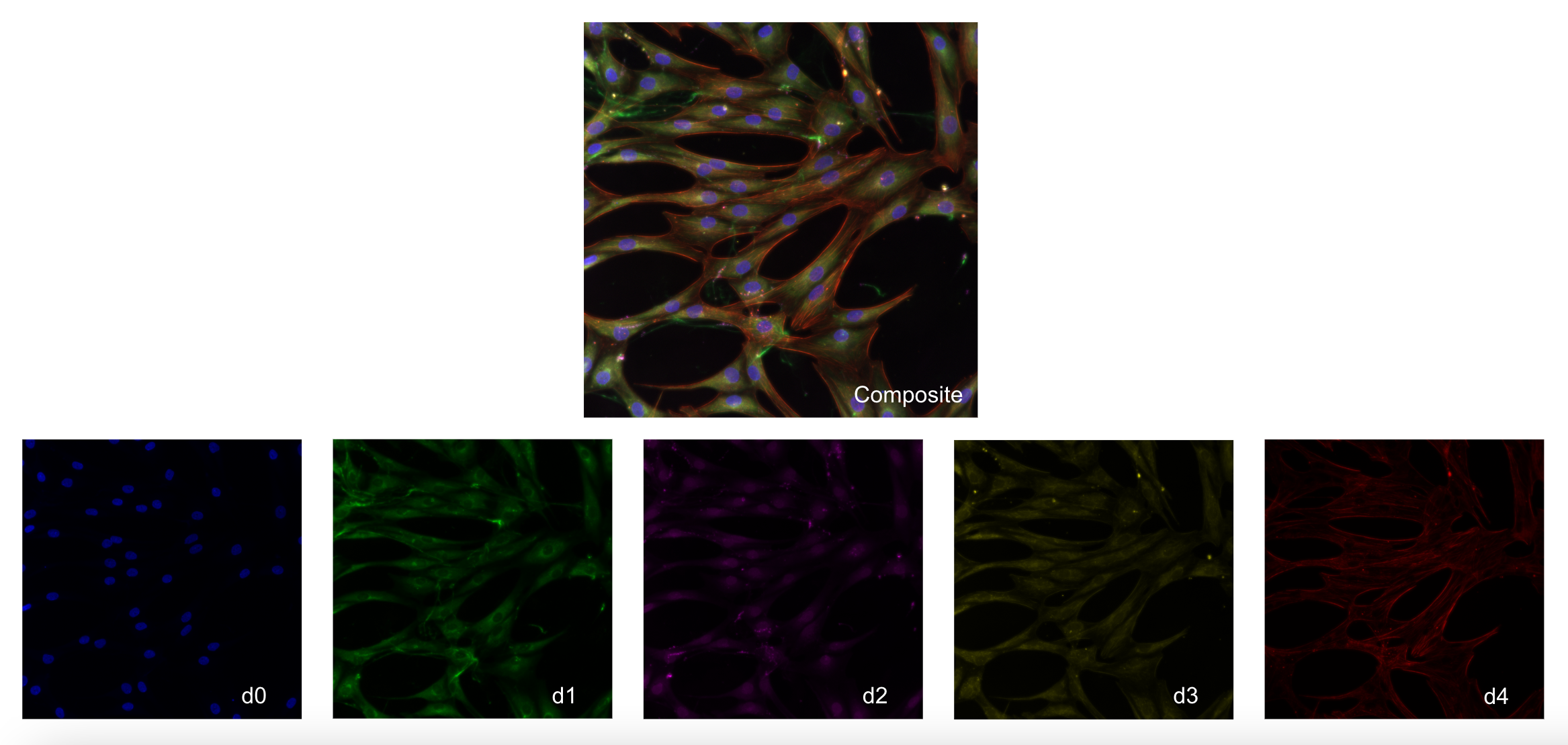
The data used in this project is a modified Cell Painting assay on cardiac fibroblasts from 3 patients that suffered from cardiac arrest.
In this modified Cell Painting, there are five channels:
d0(Nuclei)d1(Endoplasmic Reticulum)d2(Golgi/Plasma Membrane)d3(Mitochondria)d4(F-actin)
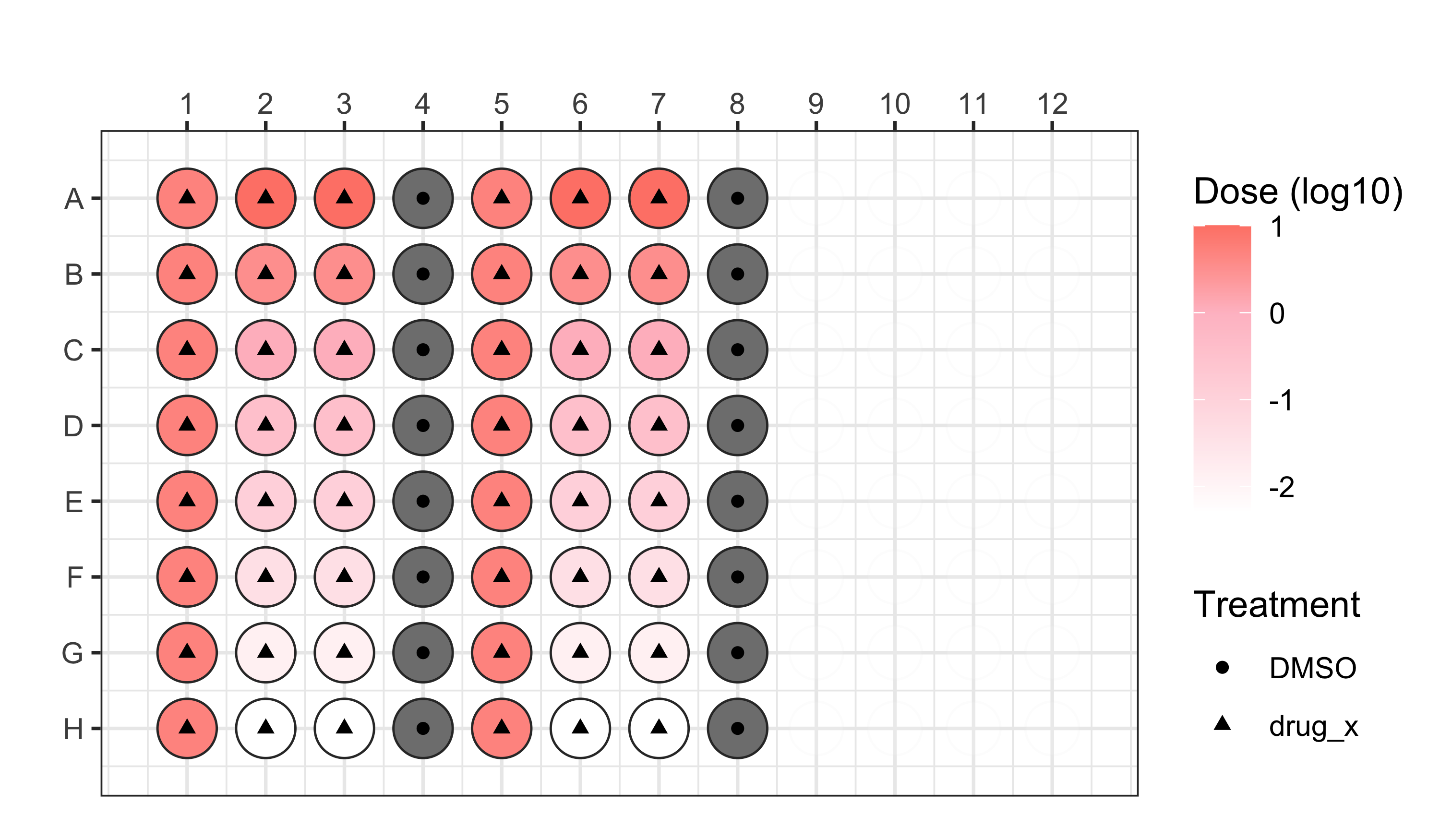
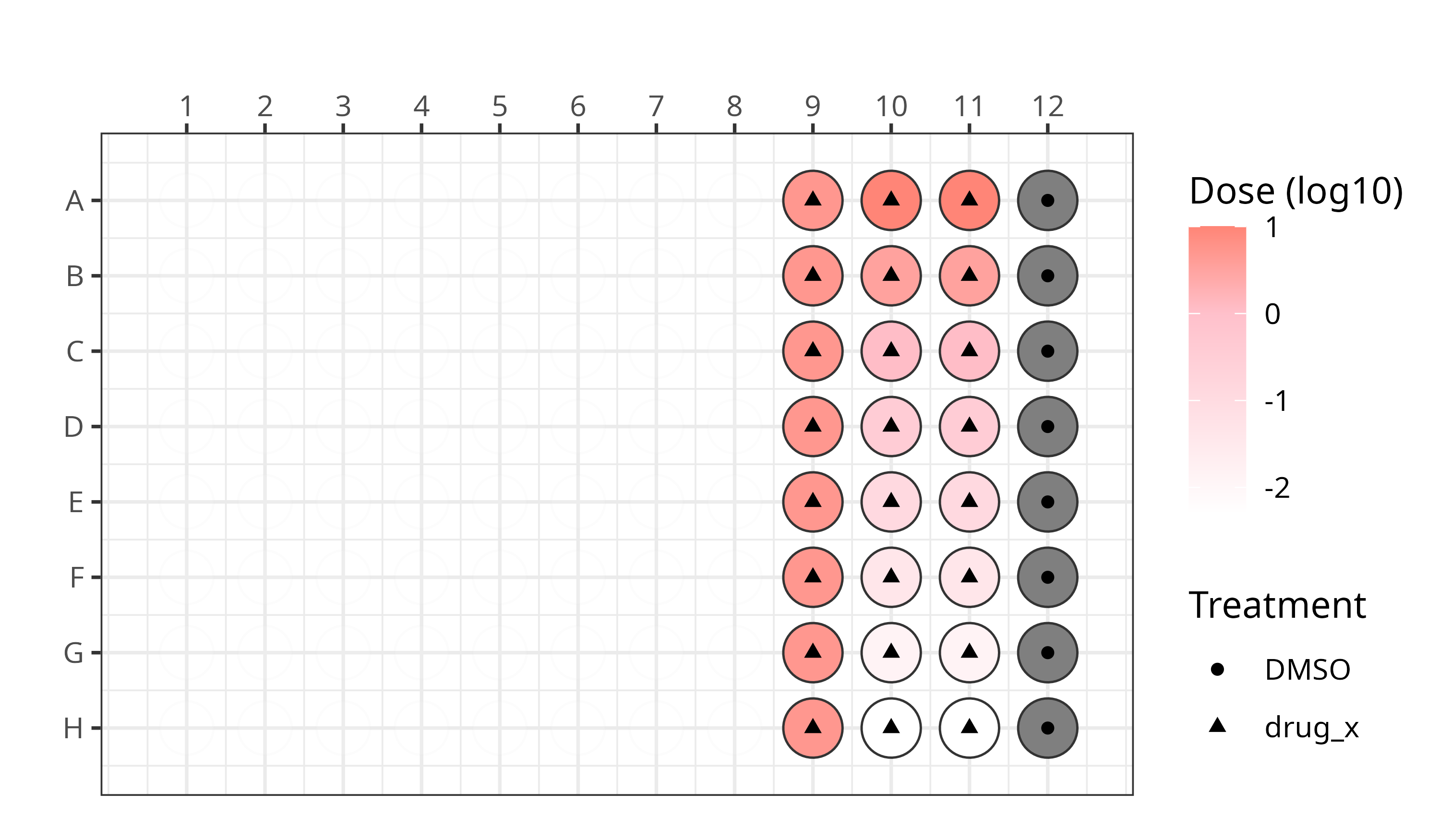
We applied this modified Cell Painting assay using the following plate design for the first two plates:
- Plate 1 = localhost220512140003_KK22-05-198
- Plate 2 = localhost220513100001_KK22-05-198_FactinAdjusted
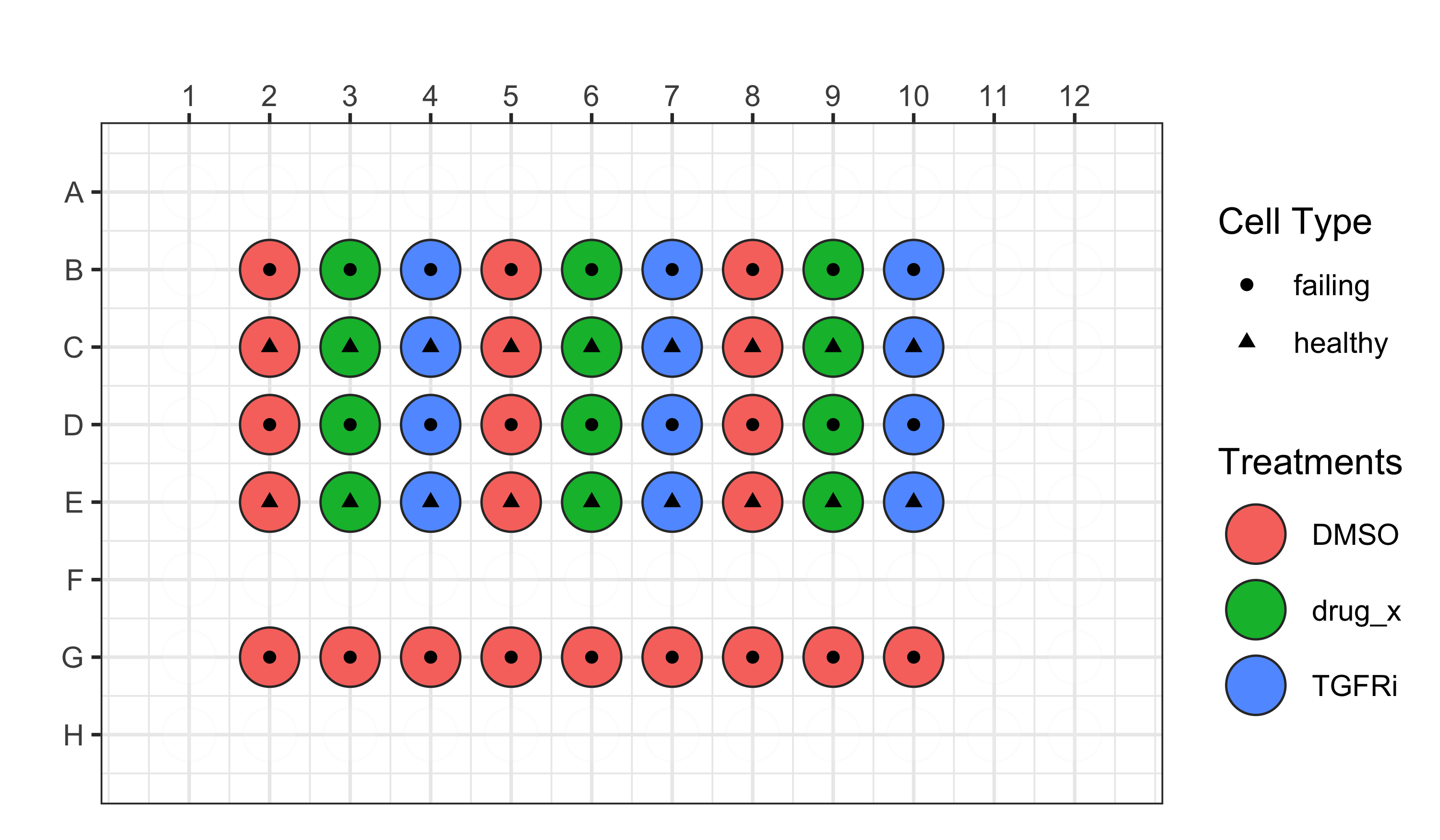
For the third plate, we are using the following plate design:
- Plate 3 = localhost230405150001
In this plate, there are only two different patients, one with a healthy heart and one that had a failing heart.
See our platemaps for more details.
The goals of this project are:
- To identify morphology features from cardiac fibroblasts that distinguish cardiac patients
- To discover a cell morphology biomarker associated with drug treatment to reverse fibrosis scarring caused by cardiac arrest.
| Module | Purpose | Description |
|---|---|---|
| 0.download_data | Download CFReT pilot data | Download pilot images for the CFReT project |
| 1.preprocessing_data | Perform Illumination Correction (IC) | Use BaSiCPy to perform IC on images per channel |
| 2.cellprofiler_processing | Apply feature extraction pipeline | Extract hundreds of morphology features per imaging channel |
| 3.process_cfret_features | Get morphology features analysis ready | Apply pycytominer to perform single cell normalization and feature selection |
| 4.analyze_data | Analyze the single cell profiles to achieve goals listed above | Several independent analyses to describe data and test hypotheses |
For all modules, we use one main environment for the repository, which includes all packages needed including installing CellProfiler v4.2.4 among other packages.
To create the environment, run the below code block:
# Run this command in terminal to create the conda environment
conda env create -f cfret_main_env.ymlMake sure that the conda environment is activated before running notebooks or scripts:
conda activate cfret_data