AceGen is a comprehensive toolkit designed to leverage reinforcement learning (RL) techniques for generative chemistry tasks, particularly in drug design. AceGen harnesses the capabilities of TorchRL, a modern library for general decision-making tasks, to provide a flexible and integrated solution for generative drug design challenges.
The full paper can be found here.
- Multiple Generative Modes: AceGen facilitates the generation of chemical libraries with different modes: de novo generation, scaffold decoration, and fragment linking.
- RL Algorithms: AceGen offers task optimization with various reinforcement learning algorithms such as Proximal Policy Optimization (PPO), Advantage Actor-Critic (A2C), Reinvent, and Augmented Hill-Climb (AHC).
- Pre-trained Models: The toolkit offers pre-trained models including Gated Recurrent Unit (GRU), Long Short-Term Memory (LSTM), and GPT-2.
- Scoring Functions : AceGen relies on MolScore, a comprehensive scoring function suite for generative chemistry, to evaluate the quality of the generated molecules.
- Customization Support: AceGen provides tutorials for integrating custom models and custom scoring functions, ensuring flexibility for advanced users.
To create the conda / mamba environment, run
conda create -n acegen python=3.10 -y
conda activate acegen
pip3 install --pre torch --index-url https://download.pytorch.org/whl/nightly/cu121
To install Tensordict, run
git clone https://github.com/pytorch/tensordict.git
cd tensordict
python setup.py install
To install TorchRL, run
git clone https://github.com/pytorch/rl.git
cd rl
python setup.py install
To install AceGen, run (use pip install -e ./ for develop mode)
pip3 install tqdm wandb hydra-core
cd acegen-open
pip install ./
Unless you intend to define your own custom scoring functions, install MolScore by running
pip3 install MolScore
To use the scaffold decoration and fragment linking, install promptsmiles by running
pip3 install promptsmiles
To learn how to configure constrained molecule generation with AcGen and promptsmiles, please refer to this tutorial.
To run the training scripts for denovo generation, run the following commands:
python scripts/a2c/a2c.py --config-name config_denovo
python scripts/ppo/ppo.py --config-name config_denovo
python scripts/reinvent/reinvent.py --config-name config_denovo
python scripts/ahc/ahc.py --config-name config_denovo
To run the training scripts for scaffold decoration, run the following commands (requires installation of promptsmiles):
python scripts/a2c/a2c.py --config-name config_scaffold
python scripts/ppo/ppo.py --config-name config_scaffold
python scripts/reinvent/reinvent.py --config-name config_scaffold
python scripts/ahc/ahc.py --config-name config_scaffold
To run the training scripts for fragment linking, run the following commands (requires installation of promptsmiles):
python scripts/a2c/a2c.py --config-name config_linking
python scripts/ppo/ppo.py --config-name config_linking
python scripts/reinvent/reinvent.py --config-name config_linking
python scripts/ahc/ahc.py --config-name config_linking
To modify training parameters, edit the corresponding YAML file in each example's directory.
Scripts are also available as executables after installation, but both the path and name of the config must be specified. For example,
ppo.py --config-path=<path_to_config_dir> --config-name=<config_name.yaml>
YAML config parameters can also be specified on the command line. For example,
ppo.py --config-path=<path_to_config_dir> --config-name=<config_name.yaml> total_smiles=100
We provide a variety of example priors that can be selected in the configuration file. These include:
-
A Gated Recurrent Unit (GRU) model
-
A Long Short-Term Memory (LSTM) model
- pre-training dataset: ChEMBL
- number of parameters: 5,807,909
-
A GPT-2 model (requires installation of HuggingFace's
transformerslibrary)- pre-training dataset: REAL 350/3 lead-like, 613.86M cpds, CXSMILES
- number of parameters: 5,030,400
To change the scoring function, adjust the molscore parameter in any configuration files. Set it to point to a valid
MolScore configuration file (e.g. ../MolScore/molscore/configs/GuacaMol/Albuterol_similarity.json).
Alternatively, you can set the molscore parameter to the name of a valid MolScore benchmark
(such as MolOpt, GuacaMol, etc.) to automatically execute each task in the benchmark. For further details on MolScore,
please refer to the MolScore repository.
Alternatively, users can define their own custom scoring functions and use them in the AceGen scripts by following the instructions in this tutorial.
We encourage users to integrate their own models into AceGen.
/acegen/models/gru.py and /acegen/models/lstm.py offer methods to create RNNs of varying sizes, which can be use
to load custom models.
Similarly, /acegen/models/gpt2.py can serve as a template for integrating HuggingFace models. A detailed guide
on integrating custom models can be found in this tutorial.
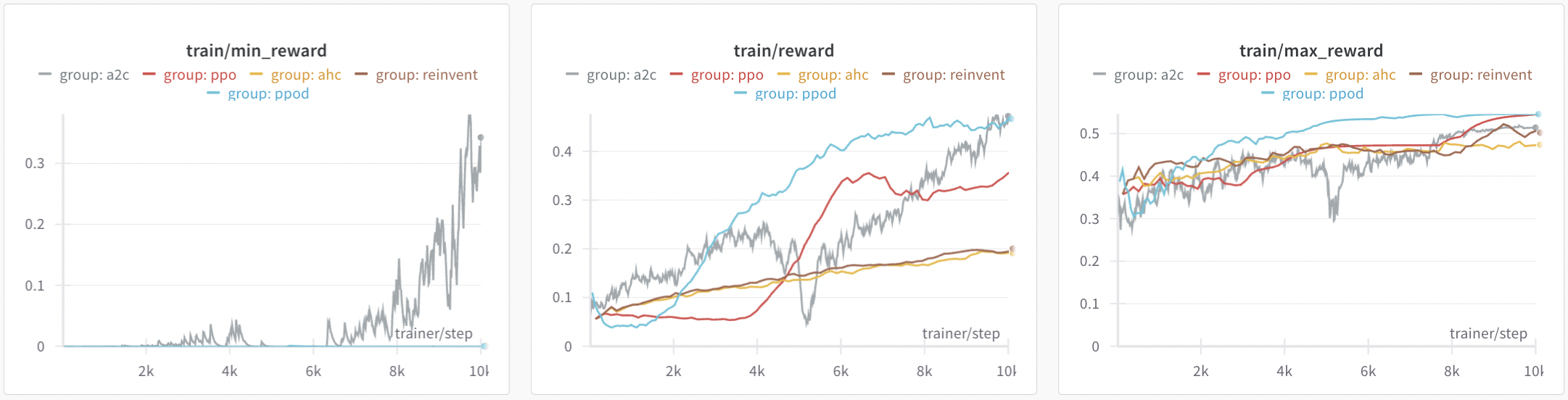
Results on the MolOpt benchmark
Algorithm comparison for the Area Under the Curve (AUC) of the top 100 molecules on MolOpt benchmark scoring functions. Each algorithm ran 5 times with different seeds, and results were averaged. We used the default configuration for each algorithm, including the GRU model for the prior. Additionally, for Reinvent we also tested the configuration proposed in the MolOpt paper.
| Task | Reinvent | Reinvent MolOpt | AHC | A2C | PPO | PPOD |
|---|---|---|---|---|---|---|
| Albuterol_similarity | 0.569 | 0.865 | 0.640 | 0.760 | 0.911 | 0.919 |
| Amlodipine_MPO | 0.506 | 0.626 | 0.505 | 0.511 | 0.553 | 0.656 |
| C7H8N2O2 | 0.615 | 0.871 | 0.563 | 0.737 | 0.864 | 0.875 |
| C9H10N2O2PF2Cl | 0.556 | 0.721 | 0.553 | 0.610 | 0.625 | 0.756 |
| Celecoxxib_rediscovery | 0.566 | 0.812 | 0.590 | 0.700 | 0.647 | 0.888 |
| Deco_hop | 0.602 | 0.657 | 0.616 | 0.605 | 0.601 | 0.646 |
| Fexofenadine_MPO | 0.668 | 0.765 | 0.680 | 0.663 | 0.687 | 0.747 |
| Median_molecules_1 | 0.199 | 0.348 | 0.197 | 0.321 | 0.362 | 0.363 |
| Median_molecules_2 | 0.195 | 0.270 | 0.208 | 0.224 | 0.236 | 0.285 |
| Mestranol_similarity | 0.454 | 0.821 | 0.514 | 0.645 | 0.728 | 0.870 |
| Osimertinib_MPO | 0.782 | 0.837 | 0.791 | 0.780 | 0.798 | 0.815 |
| Perindopril_MPO | 0.430 | 0.516 | 0.431 | 0.444 | 0.477 | 0.506 |
| QED | 0.922 | 0.931 | 0.925 | 0.927 | 0.933 | 0.933 |
| Ranolazine_MPO | 0.626 | 0.721 | 0.635 | 0.681 | 0.681 | 0.706 |
| Scaffold_hop | 0.758 | 0.834 | 0.772 | 0.764 | 0.761 | 0.808 |
| Sitagliptin_MPO | 0.226 | 0.356 | 0.219 | 0.272 | 0.295 | 0.372 |
| Thiothixene_rediscovery | 0.350 | 0.539 | 0.385 | 0.446 | 0.473 | 0.570 |
| Troglitazone_rediscovery | 0.256 | 0.447 | 0.282 | 0.305 | 0.449 | 0.511 |
| Valsartan_smarts | 0.012 | 0.014 | 0.011 | 0.010 | 0.022 | 0.022 |
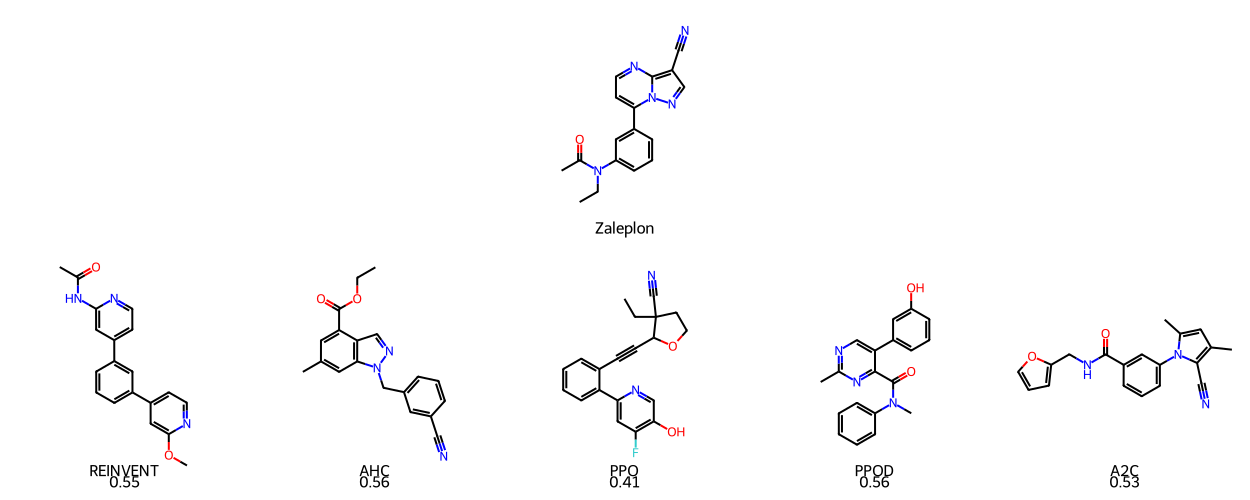
| Zaleplon_MPO | 0.408 | 0.496 | 0.412 | 0.415 | 0.469 | 0.490 |
| DRD2 | 0.907 | 0.963 | 0.906 | 0.942 | 0.967 | 0.963 |
| GSK3B | 0.738 | 0.890 | 0.719 | 0.781 | 0.863 | 0.891 |
| JNK3 | 0.640 | 0.817 | 0.649 | 0.660 | 0.770 | 0.842 |
| Total | 11.985 | 15.118 | 12.205 | 13.203 | 14.170 | 15.434 |