| title | author | date |
|---|---|---|
The Tidyverse |
Siddharth |
10-10, 2019 |
The tidyverse is a set of packages that work in harmony because they share common data representations and API design. They are designed to make it easier for data scientists to get their data "tidy", and they provide tools to assist with every step of an analysis:
| Use | Package |
|---|---|
| Data visualization | ggplot2 |
| Data manipulation | dplyr |
| Data tidying | tidyr |
| Data import | readr |
| Functional programming | purrr |
| Better data frames | tibble |
Many other packages are included as well: date/times, strings, databases, web APIs/scraping, Excel, ...
A tibble is just a data frame with some improvements:
- Printing
- Subsetting
df <- tibble(
x = runif(5),
y = letters[1:5]
)
df## # A tibble: 5 x 2
## x y
## <dbl> <chr>
## 1 0.925 a
## 2 0.301 b
## 3 0.613 c
## 4 0.563 d
## 5 0.899 e
(df2 <- as.data.frame(df))## x y
## 1 0.9245595 a
## 2 0.3006973 b
## 3 0.6126038 c
## 4 0.5632103 d
## 5 0.8988740 e
Base R is inconsistent - sometimes [ returns a data frame, sometimes a vector
class(df2[ ,1:2]) ## [1] "data.frame"
class(df2[ ,1]) ## [1] "numeric"
Subsetting a tibble with [ always returns another tibble.
class(df[ ,1:2]) ## [1] "tbl_df" "tbl" "data.frame"
class(df[ ,1])## [1] "tbl_df" "tbl" "data.frame"
To get a vector from a tibble, use $ or [[.
df[ ,'y']## # A tibble: 5 x 1
## y
## <chr>
## 1 a
## 2 b
## 3 c
## 4 d
## 5 e
df$y## [1] "a" "b" "c" "d" "e"
df[['y']]## [1] "a" "b" "c" "d" "e"
Produces useful warnings if a column doesn't exist.
df$z## Warning: Unknown or uninitialised column: 'z'.
## NULL
df2$z## NULL
Consistent import functions: read_csv(), read_tsv(), read_fwf(), etc.
read_csv('a, b, c
1, 2, 3
4, 5, 6')## # A tibble: 2 x 3
## a b c
## <int> <int> <int>
## 1 1 2 3
## 2 4 5 6
- Typically about 10x faster than base R equivalents
- They produce tibbles, don't convert characters to factors, don't screw up column names
- More reproducible
readr tries to automatically guess the type of each column based on the first 1000 rows.
challenge <- read_csv(readr_example('challenge.csv'))## Parsed with column specification:
## cols(
## x = col_integer(),
## y = col_character()
## )
## Warning in rbind(names(probs), probs_f): number of columns of result is not
## a multiple of vector length (arg 1)
## Warning: 1000 parsing failures.
## row # A tibble: 5 x 5 col row col expected actual file expected <int> <chr> <chr> <chr> <chr> actual 1 1001 x no trailing c… .2383797508… '/Library/Frameworks/R.framework… file 2 1002 x no trailing c… .4116799717… '/Library/Frameworks/R.framework… row 3 1003 x no trailing c… .7460716762… '/Library/Frameworks/R.framework… col 4 1004 x no trailing c… .7234505538… '/Library/Frameworks/R.framework… expected 5 1005 x no trailing c… .6145241374… '/Library/Frameworks/R.framework…
## ... ................. ... ........................................................................... ........ ........................................................................... ...... ........................................................................... .... ........................................................................... ... ........................................................................... ... ........................................................................... ........ ...........................................................................
## See problems(...) for more details.
Any parsing problems are not just displayed as a warning, but actually available as a tibble.
problems(challenge)## # A tibble: 1,000 x 5
## row col expected actual file
## <int> <chr> <chr> <chr> <chr>
## 1 1001 x no trailing c… .238379750… '/Library/Frameworks/R.framework…
## 2 1002 x no trailing c… .411679971… '/Library/Frameworks/R.framework…
## 3 1003 x no trailing c… .746071676… '/Library/Frameworks/R.framework…
## 4 1004 x no trailing c… .723450553… '/Library/Frameworks/R.framework…
## 5 1005 x no trailing c… .614524137… '/Library/Frameworks/R.framework…
## 6 1006 x no trailing c… .473980569… '/Library/Frameworks/R.framework…
## 7 1007 x no trailing c… .578461039… '/Library/Frameworks/R.framework…
## 8 1008 x no trailing c… .241593722… '/Library/Frameworks/R.framework…
## 9 1009 x no trailing c… .114378662… '/Library/Frameworks/R.framework…
## 10 1010 x no trailing c… .298344632… '/Library/Frameworks/R.framework…
## # … with 990 more rows
challenge <- read_csv(
readr_example("challenge.csv"),
col_types = cols(
x = col_double(),
y = col_character()
)
)
tail(challenge)## # A tibble: 6 x 2
## x y
## <dbl> <chr>
## 1 0.805 2019-11-21
## 2 0.164 2018-03-29
## 3 0.472 2014-08-04
## 4 0.718 2015-08-16
## 5 0.270 2020-02-04
## 6 0.608 2019-01-06
challenge <- read_csv(
readr_example("challenge.csv"),
col_types = cols(
x = col_double(),
y = col_date()
)
)
tail(challenge)## # A tibble: 6 x 2
## x y
## <dbl> <date>
## 1 0.805 2019-11-21
## 2 0.164 2018-03-29
## 3 0.472 2014-08-04
## 4 0.718 2015-08-16
## 5 0.270 2020-02-04
## 6 0.608 2019-01-06
It's a good idea to always explicitly specify column types!
A sample data file with 15,000 entries and 5 columns.
read_csv
init1 <- Sys.time(); df1 <- read_csv("test.csv"); final1 <- Sys.time()
final1-init1## Time difference of 0.04323101 secs
read.csv
init2 <- Sys.time(); df2 <- read.csv("test.csv"); final2 <- Sys.time()
final2-init2## Time difference of 0.1239212 secs
Writing data back to disk can be done with write_csv() or write_tsv(). These make it more likely the data can be read back in correctly.
- Strings are always encoded in UTF-8
- Dates and date-times are always stored in the same standard format (ISO8601)
Use write_excel_csv() if you plan to open a csv in Excel.
| Data type | Package |
|---|---|
| SPSS, Stata, SAS | haven |
| Excel (.xls or .xlsx) | readxl |
| Relational database | DBI + database-specific backend (RMySQL, RSQLite, ...) |
| JSON | jsonlite |
| XML | XML2 |
The dplyr package simplifies how you think about common data manipulation tasks. It provides simple "verbs", functions that correspond to these tasks, to help you take your thoughts about what you need to do and translate them into code.
| Verb | Use |
|---|---|
filter |
Filters out rows according to some conditions |
arrange |
Reorders rows according to some conditions |
select |
Selects a subset of columns |
mutate |
Adds a new column as a function of existing columns |
summarize |
Collapses a data frame to a single row |
group_by |
Breaks a data frame into groups of rows |
dplyr imports the %>% operator from the magrittr package. x %>% f(y) is equivalent to f(x, y). This allows for the verbs to be chained together in a very readable way.
Confidence intervals for the correlation from a regression model using Fisher z-transformation.
%>%:
n <- 100; rho <- 0.4
x <- rnorm(n); y <- rho*x + sqrt(1-(rho^2))*rnorm(n)
CI.r1 <- tanh( c(atanh(cor(x,y)) - 1.96/sqrt(n-3), atanh(cor(x,y)) + 1.96/sqrt(n-3) ) )
CI.r1## [1] 0.2404618 0.5671286
With %>%:
CI.r2 <- cor(x,y) %>% atanh() %>% rep(2) %>% +c(-1.96/sqrt(n-3),1.96/sqrt(n-3)) %>% tanh()
CI.r2## [1] 0.2404618 0.5671286
library(nycflights13)
flights## # A tibble: 336,776 x 19
## year month day dep_time sched_dep_time dep_delay arr_time
## <int> <int> <int> <int> <int> <dbl> <int>
## 1 2013 1 1 517 515 2 830
## 2 2013 1 1 533 529 4 850
## 3 2013 1 1 542 540 2 923
## 4 2013 1 1 544 545 -1 1004
## 5 2013 1 1 554 600 -6 812
## 6 2013 1 1 554 558 -4 740
## 7 2013 1 1 555 600 -5 913
## 8 2013 1 1 557 600 -3 709
## 9 2013 1 1 557 600 -3 838
## 10 2013 1 1 558 600 -2 753
## # … with 336,766 more rows, and 12 more variables: sched_arr_time <int>,
## # arr_delay <dbl>, carrier <chr>, flight <int>, tailnum <chr>,
## # origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>, hour <dbl>,
## # minute <dbl>, time_hour <dttm>
Suppose we want to find all the days on which both the average arrival delay and average departure delay were over 30 minutes.
Base R:
flights_avgdelay <- aggregate(flights[c('arr_delay', 'dep_delay')],
flights[c('year', 'month', 'day')],
mean, na.rm = TRUE)
flights_avgdelay30 <- flights_avgdelay[flights_avgdelay$arr_delay > 30 &
flights_avgdelay$dep_delay > 30, ]
final <- flights_avgdelay30[order(flights_avgdelay30$arr_delay,
decreasing = TRUE), ]dplyr:
final <- flights %>%
group_by(year, month, day) %>%
select(arr_delay, dep_delay) %>%
summarize(arr = mean(arr_delay, na.rm = TRUE),
dep = mean(dep_delay, na.rm = TRUE)) %>%
filter(arr > 30, dep > 30) %>%
arrange(desc(arr))## # A tibble: 36 x 5
## # Groups: year, month [10]
## year month day arr dep
## <int> <int> <int> <dbl> <dbl>
## 1 2013 3 8 85.9 83.5
## 2 2013 6 13 63.8 45.8
## 3 2013 7 22 62.8 46.7
## 4 2013 5 23 62.0 51.1
## 5 2013 7 10 59.6 52.9
## 6 2013 9 12 58.9 50.0
## 7 2013 7 1 58.3 56.2
## 8 2013 12 17 55.9 40.7
## 9 2013 8 8 55.5 43.3
## 10 2013 12 5 51.7 52.3
## # … with 26 more rows
Each of these datasets shows the same information: country, year, population, and cases.
table1## # A tibble: 6 x 4
## country year cases population
## <chr> <int> <int> <int>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583
table2## # A tibble: 12 x 4
## country year type count
## <chr> <int> <chr> <int>
## 1 Afghanistan 1999 cases 745
## 2 Afghanistan 1999 population 19987071
## 3 Afghanistan 2000 cases 2666
## 4 Afghanistan 2000 population 20595360
## 5 Brazil 1999 cases 37737
## 6 Brazil 1999 population 172006362
## 7 Brazil 2000 cases 80488
## 8 Brazil 2000 population 174504898
## 9 China 1999 cases 212258
## 10 China 1999 population 1272915272
## 11 China 2000 cases 213766
## 12 China 2000 population 1280428583
table3## # A tibble: 6 x 3
## country year rate
## * <chr> <int> <chr>
## 1 Afghanistan 1999 745/19987071
## 2 Afghanistan 2000 2666/20595360
## 3 Brazil 1999 37737/172006362
## 4 Brazil 2000 80488/174504898
## 5 China 1999 212258/1272915272
## 6 China 2000 213766/1280428583
table4a## # A tibble: 3 x 3
## country `1999` `2000`
## * <chr> <int> <int>
## 1 Afghanistan 745 2666
## 2 Brazil 37737 80488
## 3 China 212258 213766
table4b## # A tibble: 3 x 3
## country `1999` `2000`
## * <chr> <int> <int>
## 1 Afghanistan 19987071 20595360
## 2 Brazil 172006362 174504898
## 3 China 1272915272 1280428583
- Each variable must have its own column.
- Each observation must have its own row.
- Each value must have its own cell.
Practical instructions:
- Put each dataset in a tibble.
- Put each variable in a column.
Which one of those previous tables is tidy?
Two main advantages:
- Good to pick one consistent way of storing data.
- Ensuring each variable has a column lets you take full advantage of R's vectorization.
All of the tidyverse packages are designed to work with tidy data.
table1 %>%
mutate(rate = cases / population * 10000)## # A tibble: 6 x 5
## country year cases population rate
## <chr> <int> <int> <int> <dbl>
## 1 Afghanistan 1999 745 19987071 0.373
## 2 Afghanistan 2000 2666 20595360 1.29
## 3 Brazil 1999 37737 172006362 2.19
## 4 Brazil 2000 80488 174504898 4.61
## 5 China 1999 212258 1272915272 1.67
## 6 China 2000 213766 1280428583 1.67
Data is often organized to make entry as easy as possible (e.g. in Excel). Often there will be one of two problems:
- One variable is spread across multiple columns.
- One observation is scattered across multiple rows.
To fix these, we can use spread() and gather() from the tidyr package.
table4a## # A tibble: 3 x 3
## country `1999` `2000`
## * <chr> <int> <int>
## 1 Afghanistan 745 2666
## 2 Brazil 37737 80488
## 3 China 212258 213766
The 1999 and 2000 columns are values, not variables! The correct variable would be "year". To gather those columns, we need to specify:
- The set of columns that represent values:
1999and2000 - The name of the variable whose values are the column names:
year - The name of the variable whose values are in the cells:
cases
table4a %>%
gather(`1999`, `2000`, key = 'year', value = 'cases')## # A tibble: 6 x 3
## country year cases
## <chr> <chr> <int>
## 1 Afghanistan 1999 745
## 2 Brazil 1999 37737
## 3 China 1999 212258
## 4 Afghanistan 2000 2666
## 5 Brazil 2000 80488
## 6 China 2000 213766
table2## # A tibble: 12 x 4
## country year type count
## <chr> <int> <chr> <int>
## 1 Afghanistan 1999 cases 745
## 2 Afghanistan 1999 population 19987071
## 3 Afghanistan 2000 cases 2666
## 4 Afghanistan 2000 population 20595360
## 5 Brazil 1999 cases 37737
## 6 Brazil 1999 population 172006362
## 7 Brazil 2000 cases 80488
## 8 Brazil 2000 population 174504898
## 9 China 1999 cases 212258
## 10 China 1999 population 1272915272
## 11 China 2000 cases 213766
## 12 China 2000 population 1280428583
Here one observation is scattered over two rows. The type column actually contains two variables, cases and population. To tidy this, we need to specify:
- The column that contains variable names:
type - The column that contains values from multiple variables:
count
table2 %>%
spread(key = type, value = count)## # A tibble: 6 x 4
## country year cases population
## <chr> <int> <int> <int>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583
How do we fix this?
table3## # A tibble: 6 x 3
## country year rate
## * <chr> <int> <chr>
## 1 Afghanistan 1999 745/19987071
## 2 Afghanistan 2000 2666/20595360
## 3 Brazil 1999 37737/172006362
## 4 Brazil 2000 80488/174504898
## 5 China 1999 212258/1272915272
## 6 China 2000 213766/1280428583
The rate column contains two variables: cases and population. We can separate it into two columns with the separate() function.
table3 %>%
separate(rate, into = c('cases', 'population'), sep = '/', convert = TRUE)## # A tibble: 6 x 4
## country year cases population
## <chr> <int> <int> <int>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583
By default, separate() will split on any non-alphanumeric character. The convert argument will try to convert the resulting columns into a better type.
You can do the inverse operation with unite(), but it isn't all that useful.
tidyr::who## # A tibble: 7,240 x 60
## country iso2 iso3 year new_sp_m014 new_sp_m1524 new_sp_m2534
## <chr> <chr> <chr> <int> <int> <int> <int>
## 1 Afghan… AF AFG 1980 NA NA NA
## 2 Afghan… AF AFG 1981 NA NA NA
## 3 Afghan… AF AFG 1982 NA NA NA
## 4 Afghan… AF AFG 1983 NA NA NA
## 5 Afghan… AF AFG 1984 NA NA NA
## 6 Afghan… AF AFG 1985 NA NA NA
## 7 Afghan… AF AFG 1986 NA NA NA
## 8 Afghan… AF AFG 1987 NA NA NA
## 9 Afghan… AF AFG 1988 NA NA NA
## 10 Afghan… AF AFG 1989 NA NA NA
## # … with 7,230 more rows, and 53 more variables: new_sp_m3544 <int>,
## # new_sp_m4554 <int>, new_sp_m5564 <int>, new_sp_m65 <int>,
## # new_sp_f014 <int>, new_sp_f1524 <int>, new_sp_f2534 <int>,
## # new_sp_f3544 <int>, new_sp_f4554 <int>, new_sp_f5564 <int>,
## # new_sp_f65 <int>, new_sn_m014 <int>, new_sn_m1524 <int>,
## # new_sn_m2534 <int>, new_sn_m3544 <int>, new_sn_m4554 <int>,
## # new_sn_m5564 <int>, new_sn_m65 <int>, new_sn_f014 <int>,
## # new_sn_f1524 <int>, new_sn_f2534 <int>, new_sn_f3544 <int>,
## # new_sn_f4554 <int>, new_sn_f5564 <int>, new_sn_f65 <int>,
## # new_ep_m014 <int>, new_ep_m1524 <int>, new_ep_m2534 <int>,
## # new_ep_m3544 <int>, new_ep_m4554 <int>, new_ep_m5564 <int>,
## # new_ep_m65 <int>, new_ep_f014 <int>, new_ep_f1524 <int>,
## # new_ep_f2534 <int>, new_ep_f3544 <int>, new_ep_f4554 <int>,
## # new_ep_f5564 <int>, new_ep_f65 <int>, newrel_m014 <int>,
## # newrel_m1524 <int>, newrel_m2534 <int>, newrel_m3544 <int>,
## # newrel_m4554 <int>, newrel_m5564 <int>, newrel_m65 <int>,
## # newrel_f014 <int>, newrel_f1524 <int>, newrel_f2534 <int>,
## # newrel_f3544 <int>, newrel_f4554 <int>, newrel_f5564 <int>,
## # newrel_f65 <int>
"While model inputs usually require tidy inputs, such attention to detail doesn't carry over to model outputs. Outputs such as predictions and estimated coefficients aren't always tidy. This makes it more difficult to combine results from multiple models."
fit <- lm(mpg ~ wt, data = mtcars)
fit##
## Call:
## lm(formula = mpg ~ wt, data = mtcars)
##
## Coefficients:
## (Intercept) wt
## 37.285 -5.344
summary(fit)##
## Call:
## lm(formula = mpg ~ wt, data = mtcars)
##
## Residuals:
## Min 1Q Median 3Q Max
## -4.5432 -2.3647 -0.1252 1.4096 6.8727
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 37.2851 1.8776 19.858 < 2e-16 ***
## wt -5.3445 0.5591 -9.559 1.29e-10 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 3.046 on 30 degrees of freedom
## Multiple R-squared: 0.7528, Adjusted R-squared: 0.7446
## F-statistic: 91.38 on 1 and 30 DF, p-value: 1.294e-10
Most models produce output at three levels:
- Model level:
$R^2$ , residual standard error, MSE - Term level: coefficient estimates, p-values, per-cluster information
- Observation level: predictions, residuals, cluster assignments
broom provides functions to tidy model output in each of these three ways.
glance(): constructs a one-row summary of the model.tidy(): summarizes the model's findings in a data frame.augment(): adds columns to the original data.
This is extremely powerful when combined with dplyr.
library(broom)
glance(fit) %>% as_tibble()## # A tibble: 1 x 11
## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
## 1 0.753 0.745 3.05 91.4 1.29e-10 2 -80.0 166. 170.
## # … with 2 more variables: deviance <dbl>, df.residual <int>
tidy(fit) %>% as_tibble()## # A tibble: 2 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 37.3 1.88 19.9 8.24e-19
## 2 wt -5.34 0.559 -9.56 1.29e-10
augment(fit) %>% as_tibble()## # A tibble: 32 x 10
## .rownames mpg wt .fitted .se.fit .resid .hat .sigma .cooksd
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Mazda RX4 21 2.62 23.3 0.634 -2.28 0.0433 3.07 1.33e-2
## 2 Mazda RX… 21 2.88 21.9 0.571 -0.920 0.0352 3.09 1.72e-3
## 3 Datsun 7… 22.8 2.32 24.9 0.736 -2.09 0.0584 3.07 1.54e-2
## 4 Hornet 4… 21.4 3.22 20.1 0.538 1.30 0.0313 3.09 3.02e-3
## 5 Hornet S… 18.7 3.44 18.9 0.553 -0.200 0.0329 3.10 7.60e-5
## 6 Valiant 18.1 3.46 18.8 0.555 -0.693 0.0332 3.10 9.21e-4
## 7 Duster 3… 14.3 3.57 18.2 0.573 -3.91 0.0354 3.01 3.13e-2
## 8 Merc 240D 24.4 3.19 20.2 0.539 4.16 0.0313 3.00 3.11e-2
## 9 Merc 230 22.8 3.15 20.5 0.540 2.35 0.0314 3.07 9.96e-3
## 10 Merc 280 19.2 3.44 18.9 0.553 0.300 0.0329 3.10 1.71e-4
## # … with 22 more rows, and 1 more variable: .std.resid <dbl>
What if we wanted to fit a separate linear model to different groups of data?
regressions <- mtcars %>%
group_by(cyl) %>%
do(fit = lm(mpg ~ wt + qsec + gear, data = .))
regressions## Source: local data frame [3 x 2]
## Groups: <by row>
##
## # A tibble: 3 x 2
## cyl fit
## * <dbl> <list>
## 1 4 <S3: lm>
## 2 6 <S3: lm>
## 3 8 <S3: lm>
Now we can use the tidying functions from broom on the fit column.
regressions %>% glance(fit)## # A tibble: 3 x 12
## # Groups: cyl [3]
## cyl r.squared adj.r.squared sigma statistic p.value df logLik AIC
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl>
## 1 4 0.671 0.530 3.09 4.76 0.0410 4 -25.5 61.1
## 2 6 0.819 0.638 0.874 4.53 0.123 4 -6.02 22.0
## 3 8 0.509 0.362 2.05 3.45 0.0593 4 -27.5 65.1
## # … with 3 more variables: BIC <dbl>, deviance <dbl>, df.residual <int>
regressions %>% tidy(fit)## # A tibble: 12 x 6
## # Groups: cyl [3]
## cyl term estimate std.error statistic p.value
## <dbl> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 4 (Intercept) 0.930 21.8 0.0427 0.967
## 2 4 wt -7.67 2.23 -3.44 0.0109
## 3 4 qsec 1.62 0.892 1.81 0.113
## 4 4 gear 3.02 2.29 1.32 0.230
## 5 6 (Intercept) 83.0 29.3 2.84 0.0658
## 6 6 wt 4.34 5.21 0.833 0.466
## 7 6 qsec -3.09 1.90 -1.63 0.202
## 8 6 gear -5.51 2.78 -1.98 0.142
## 9 8 (Intercept) 11.1 16.8 0.662 0.523
## 10 8 wt -2.84 0.894 -3.18 0.00989
## 11 8 qsec 0.866 0.861 1.00 0.339
## 12 8 gear 0.255 1.28 0.200 0.846
regressions %>% augment(fit)## # A tibble: 32 x 12
## # Groups: cyl [3]
## cyl mpg wt qsec gear .fitted .se.fit .resid .hat .sigma
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 4 22.8 2.32 18.6 4 25.3 1.13 -2.47 0.135 3.16
## 2 4 24.4 3.19 20 4 20.8 1.92 3.55 0.386 2.78
## 3 4 22.8 3.15 22.9 4 25.8 2.80 -3.04 0.821 1.60
## 4 4 32.4 2.2 19.5 4 27.6 1.02 4.82 0.108 2.61
## 5 4 30.4 1.62 18.5 4 30.5 1.62 -0.137 0.273 3.34
## 6 4 33.9 1.84 19.9 4 31.1 1.73 2.82 0.313 3.04
## 7 4 21.5 2.46 20.0 3 23.4 2.36 -1.91 0.583 3.11
## 8 4 27.3 1.94 18.9 4 28.7 1.19 -1.40 0.148 3.28
## 9 4 26 2.14 16.7 5 26.6 2.12 -0.587 0.469 3.32
## 10 4 30.4 1.51 16.9 5 31.7 2.05 -1.32 0.441 3.26
## # … with 22 more rows, and 2 more variables: .cooksd <dbl>,
## # .std.resid <dbl>
Consider the mtcars dataset for variable selection.
df <- mtcars %>% as_tibble()
vars <- c("wt","qsec", "gear","hp")
id <- c(1:4) %>% lapply(function(i)combn(1:4,i,simplify=F)) %>% unlist(,recursive=F)
Formulas <- id %>% sapply(function(i)paste("mpg~",paste(vars[i],collapse = "+")))
models <- Formulas %>% lapply(function(i) lm(as.formula(i),df))
Formulas## [1] "mpg~ wt" "mpg~ qsec" "mpg~ gear"
## [4] "mpg~ hp" "mpg~ wt+qsec" "mpg~ wt+gear"
## [7] "mpg~ wt+hp" "mpg~ qsec+gear" "mpg~ qsec+hp"
## [10] "mpg~ gear+hp" "mpg~ wt+qsec+gear" "mpg~ wt+qsec+hp"
## [13] "mpg~ wt+gear+hp" "mpg~ qsec+gear+hp" "mpg~ wt+qsec+gear+hp"
models[[11]]##
## Call:
## lm(formula = as.formula(i), data = df)
##
## Coefficients:
## (Intercept) wt qsec gear
## 12.9432 -4.6178 1.0486 0.8914
summaries <- models %>% sapply(function(i) glance(summary(i))) %>% t()
rownames(summaries) <- Formulassummaries## adj.r.squared sigma statistic p.value df
## mpg~ wt 0.7445939 3.045882 91.37533 1.293959e-10 2
## mpg~ qsec 0.1478062 5.563738 6.376702 0.01708199 2
## mpg~ gear 0.2050292 5.373695 8.995144 0.005400948 2
## mpg~ hp 0.5891853 3.862962 45.4598 1.787835e-07 2
## mpg~ wt+qsec 0.8144448 2.596175 69.03311 9.394765e-12 3
## mpg~ wt+gear 0.7368661 3.091618 44.40536 1.48796e-09 3
## mpg~ wt+hp 0.8148396 2.593412 69.21121 9.109054e-12 3
## mpg~ qsec+gear 0.4813285 4.340539 15.38404 2.792324e-05 3
## mpg~ qsec+hp 0.6118339 3.754968 25.43136 4.176204e-07 3
## mpg~ gear+hp 0.734107 3.107785 43.79412 1.730921e-09 3
## mpg~ wt+qsec+gear 0.8151711 2.591089 46.57422 5.200213e-11 4
## mpg~ wt+qsec+hp 0.8170643 2.577785 47.15282 4.506411e-11 4
## mpg~ wt+gear+hp 0.8175771 2.574169 47.31161 4.333845e-11 4
## mpg~ qsec+gear+hp 0.7251222 3.159856 28.25913 1.284466e-08 4
## mpg~ wt+qsec+gear+hp 0.8220952 2.542092 36.81264 1.449667e-10 5
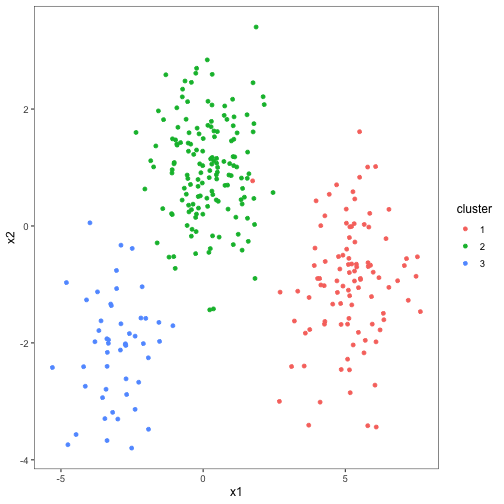
set.seed(2014)
centers <- data_frame(cluster = factor(1:3),
size = c(100, 150, 50),
x1 = c(5, 0, -3),
x2 = c(-1, 1, -2))
points <- centers %>%
group_by(cluster) %>%
do(data_frame(x1 = rnorm(.$size[1], .$x1[1]),
x2 = rnorm(.$size[1], .$x2[1])))
ggplot(points, aes(x1, x2, color = cluster)) + geom_point()## Warning: `data_frame()` is deprecated, use `tibble()`.
## This warning is displayed once per session.
Let's try doing
pts_matrix <- cbind(x1 = points$x1, x2 = points$x2)
kclust <- kmeans(pts_matrix, 3)
kclust## K-means clustering with 3 clusters of sizes 99, 151, 50
##
## Cluster means:
## x1 x2
## 1 5.1791917 -0.9865170
## 2 0.1583797 0.9797098
## 3 -3.0653196 -2.0887225
##
## Clustering vector:
## [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [36] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [71] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2
## [106] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [141] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [176] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [211] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [246] 2 2 2 2 2 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3
## [281] 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3
##
## Within cluster sum of squares by cluster:
## [1] 204.96483 257.20360 78.64255
## (between_SS / total_SS = 85.1 %)
##
## Available components:
##
## [1] "cluster" "centers" "totss" "withinss"
## [5] "tot.withinss" "betweenss" "size" "iter"
## [9] "ifault"
kclust %>% augment(pts_matrix) %>% as_tibble()## # A tibble: 300 x 3
## x1 x2 .cluster
## <dbl> <dbl> <fct>
## 1 4.43 0.542 1
## 2 5.32 -0.941 1
## 3 5.13 -1.58 1
## 4 6.35 -1.60 1
## 5 3.71 -3.41 1
## 6 5.32 -0.772 1
## 7 5.27 0.586 1
## 8 5.40 -0.701 1
## 9 5.46 -1.82 1
## 10 7.15 -0.559 1
## # … with 290 more rows
tidy(kclust) %>% as_tibble()## # A tibble: 3 x 5
## x1 x2 size withinss cluster
## <dbl> <dbl> <int> <dbl> <fct>
## 1 5.18 -0.987 99 205. 1
## 2 0.158 0.980 151 257. 2
## 3 -3.07 -2.09 50 78.6 3
Let's say we wanted to look at different choices for kmeans once for every value of
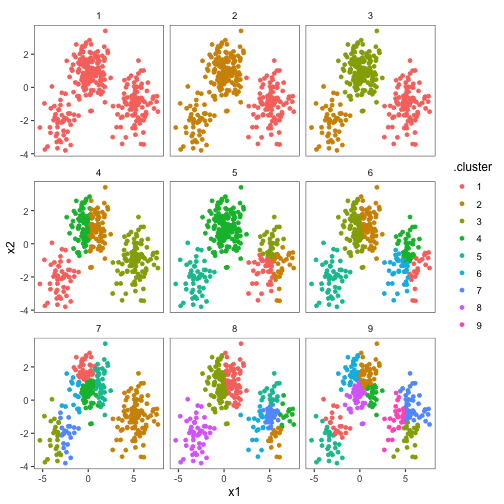
kclusts <- data_frame(k = 1:9) %>% group_by(k) %>% do(kclust = kmeans(pts_matrix, .$k))Then tidy the results in all three ways.
clusters <- kclusts %>% group_by(k) %>% do(tidy(.$kclust[[1]]))
assignments <- kclusts %>% group_by(k) %>% do(augment(.$kclust[[1]], pts_matrix))
clusterings <- kclusts %>% group_by(k) %>% do(glance(.$kclust[[1]]))
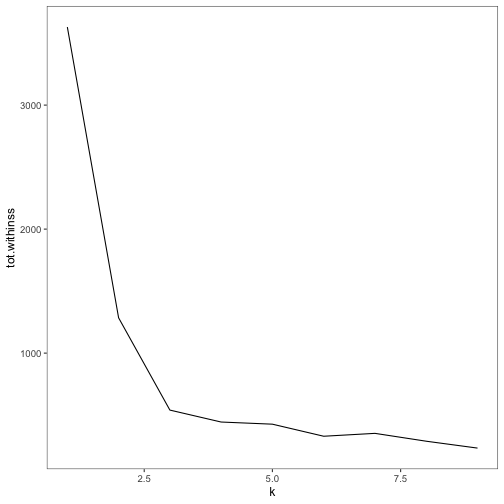
clusterings## # A tibble: 9 x 5
## # Groups: k [9]
## k totss tot.withinss betweenss iter
## <int> <dbl> <dbl> <dbl> <int>
## 1 1 3630. 3630. -3.64e-12 1
## 2 2 3630. 1285. 2.34e+ 3 1
## 3 3 3630. 541. 3.09e+ 3 2
## 4 4 3630. 444. 3.19e+ 3 3
## 5 5 3630. 427. 3.20e+ 3 3
## 6 6 3630. 330. 3.30e+ 3 3
## 7 7 3630. 353. 3.28e+ 3 2
## 8 8 3630. 290. 3.34e+ 3 3
## 9 9 3630. 234. 3.40e+ 3 4
Now we can plot the data colored by cluster.
p1 <- ggplot(assignments, aes(x1, x2)) + geom_point(aes(color = .cluster)) + facet_wrap(~k)
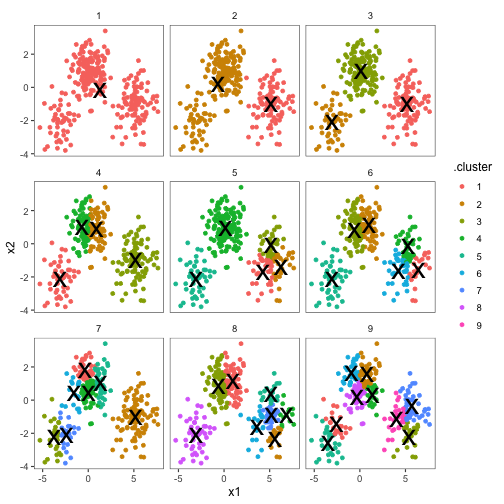
p1We can use the tidy() output to add the centers.
p2 <- p1 + geom_point(data = clusters, size = 10, shape = 'x')
p2Finally, we can use the glance() output to see the total within sum of squares for each
ggplot(clusterings, aes(k, tot.withinss)) + geom_line()